P09-08
Evaluation of anticancer activity and investigation of cellular uptake mechanism of drug-loaded DNA Origami dendrimers for application to drug delivery system
Koichi TANIMOTO *, Yuki MINAMIDE, Maria HASHIMOTO, Haruki TANAKA, Yuki MANO, Akinori KUZUYA
Department of Chemistry and Materials Engineering, Kansai University
( * E-mail: k769965@kansai-u.ac.jp )
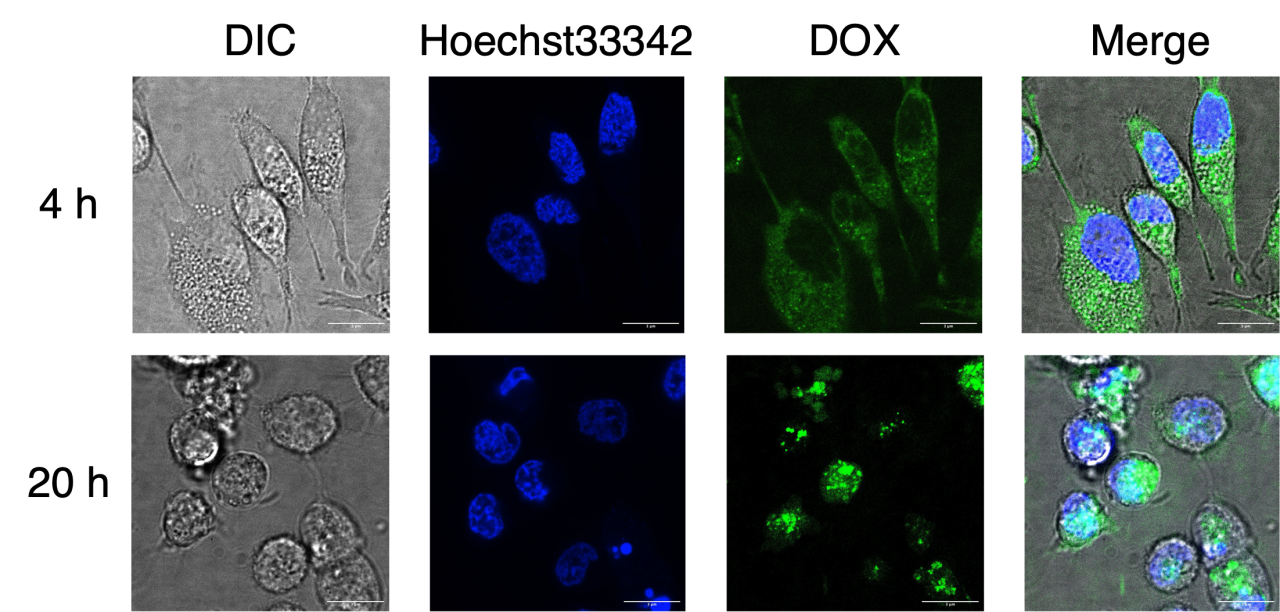
In recent years, drug delivery systems (DDS) have been actively developed as a technology for efficient and safe delivery of drugs to the site of disease. Nanocarriers are used to deliver therapeutic agents to target cells in DDS. One of the possible nanocarriers is DNA Origami, which can create arbitrary structures by folding long single-stranded circular DNA with many short single-stranded DNAs [1]. We have developed "DNA Origami dendrimers," which have four generations of four-branched dendritic structure from the center. This structure can bind doxorubicin (DOX), a popular anticancer drug, to the stems.
In this study, we delivered DOX-loaded DNA Origami dendrimers into human cervical carcinoma (HeLa) cells and evaluated anticancer activity and cellular uptake mechanism.
As a result, we confirmed that red fluorescence from Ethidium Homodimer-1, which stains dead cells, was not observed after 4 hours of incubation in the Live/Dead Assay, but many cancer cells were dead after 20 hours. This may be due to the gradual release of DOX from DNA Origami dendrimers over time. This is also supported quantitatively by MTT Assay. In investigation of the cellular uptake mechanism, it was confirmed that green fluorescence from FAM modified with DNA Origami dendrimers co-localized with red fluorescence from LysoTracker Red DND-99, which stains lysosomes around the cell nucleus. This suggests that DNA Origami dendrimers were internalized via the endocytosis pathway. Similar results were obtained with DOX-loaded DNA Origami dendrimers, showing cellular uptake via the endocytosis pathway, and then only DOX was released from lysosomes and transported to the nucleus.