P09-02
Size-Selective Capturing of Exosomes Using DNA Tripods
Ryosuke IINUMA *1, 2, Xiaoxia CHEN3, Takeya MASUBUCHI2, 4, Takuya UEDA2, 5, Hisashi TADAKUMA2, 3, 6
1JSR Life Sciences Corporation
2Graduate School of Frontier Science, The University of Tokyo
3School of Life Science and Technology, ShanghaiTech University
4Department of Cell and Developmental Biology, School of Biological Sciences, University of California San Diego
5Graduate School of Science and Engineering, Waseda University
6Gene Editing Center, ShanghaiTech University
( * E-mail: Ryousuke_Iinuma@jls.jsr.co.jp )
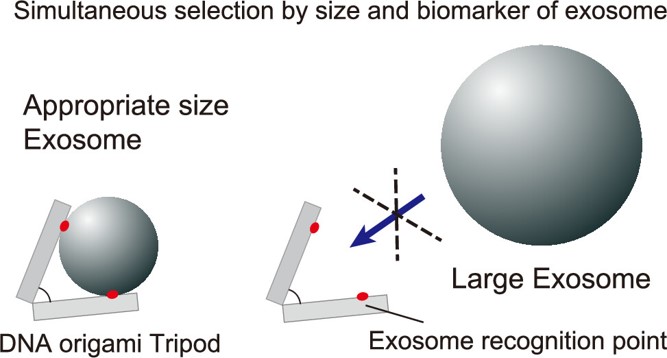
Fractionating and characterizing target samples are fundamental to the analysis of biomolecules. Extracellular vesicles (EVs), containing information regarding the cellular birthplace, are promising targets for biology and medicine. However, the requirement for multiple-step purification in conventional methods hinders analysis of small samples. Here, we apply a DNA origami tripod with a defined aperture of binders (e.g., antibodies against EV biomarkers), which allows us to capture the target molecule.
For the first step, we verify the concept of size selective capture of target molecules using magnetic beads as the model, and confirm that the DNA tripod only captures beads whose size fits inside the defined aperture. Similarly, using exosomes from HT-29 cell line as a model, we show that our tripod nanodevice can capture a specific size range of EVs with cognate biomarkers from a broad distribution of crude EV mixtures. Next, we demonstrate that the size of captured EVs can be controlled by changing the aperture of the tripods. Finally, we applied the principle to a solid-supported capture system for EVs. This simultaneous selection with the size and biomarker approach should simplify the EV purification process and contribute to the precise analysis of target biomolecules from small samples.