P07-33
Development of an efficient compound 3D conformer search system based on relative position of fragments
Tomoya SAITO *, Keisuke YANAGISAWA, Yutaka AKIYAMA
Department of Computer Science , Institute of Science Tokyo
( * E-mail: t_saito@bi.c.titech.ac.jp )
Fragment-based docking has recently been investigated as a mean to speed up large-scale docking calculations. In the context of docking calculations, a fragment is defined as a substructure of a compound that has no internal degrees of freedom. Fragment-based docking estimates the docking score and binding pose of the entire compound by combining the results of fragment-wise docking calculations. It uses fragments as the elements of a compound, and since these fragments are common to other compounds, the results of the fragment-wise calculation can be reused. However, when reconstructing the feasible conformations of a compound from fragments, it is necessary to take into account collisions and possible covalent bonding between fragments, which is a bottleneck in fragment-based docking. For example, eHiTS [1] reconstructs it by solving the NP-hard maximum clique finding problem.
To address the issue, we propose fragment-based conformer database and search system rather than conformer reconstruction algorithms. This system retrieves adequate conformations, thus avoiding the need to consider spatial collisions.
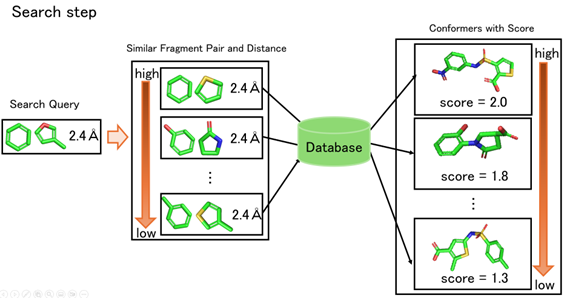
In the database preparation step, pre-generated 3D conformers are decomposed into fragments with positions, and all pairs of these fragments with Euclidean distance information are registered in the database. In the search step, users input a query consisted of two fragments with distance constraints between them, and the database outputs registered conformers which match the query with a reasonable error margin.
For highly accurate and precise search, it is ideal to distinguish all fragment types. However, with hundreds of thousands of fragment types, it is not practical to record every possible fragment pair. To solve this problem, we introduced a similarity search method that clusters similar fragments as a group. This approach allows us to reduce the number of fragment types in the database and enhance recall. Additionally, we are working on improving precision by ranking the search results based on the fragment similarities.
[1] Zsolt Zsoldos, Darryl Reid, Aniko Simon, Sayyed Bashir Sadjad, A.Peter Johnson, Journal of Molecular Graphics and Modelling, 198-212, 2007