P07-32
Quantitative Assessment of Protein−Ligand Activity Prediction from 3D Docking Poses for Urate Transporter 1
MARTIN *1, Mochammad Arfin Fardiansyah NASUTION2, Ziwei ZHOU2, Xingran WANG2, Reiko WATANABE2, Kenji MIZUGUCHI2, Ichigaku TAKIGAWA1, 3
1WPI-ICReDD, Hokkaido University
2Institute of Protein Research, Osaka University
3Institute for Liberal Arts and Sciences, Kyoto University
( * E-mail: martin@icredd.hokudai.ac.jp )
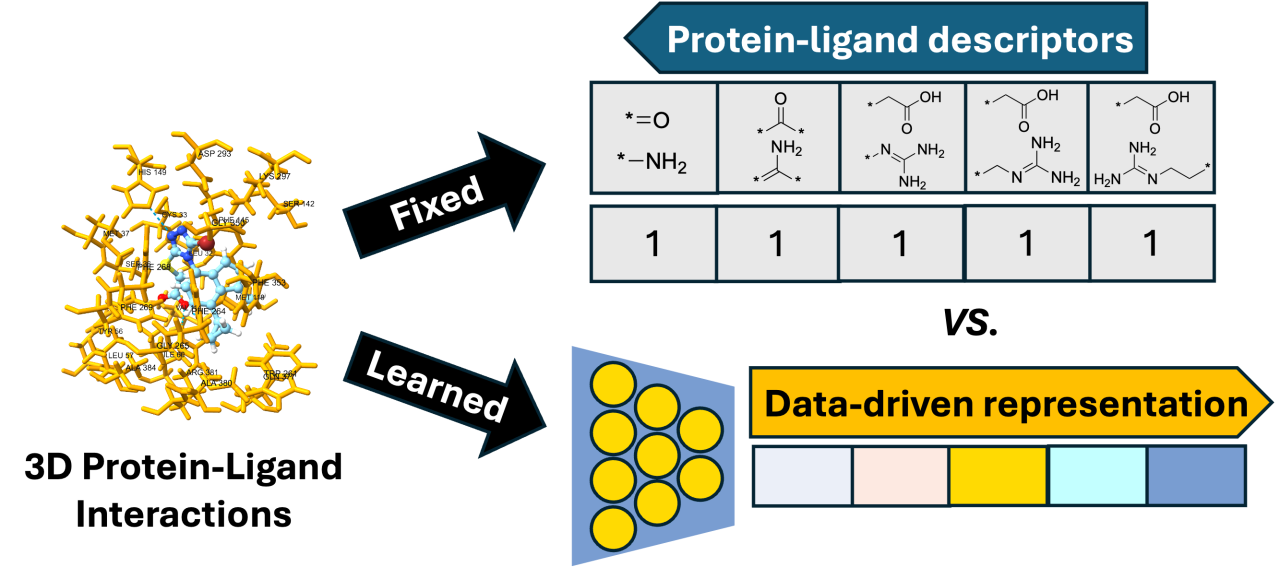
Urate transporter 1 (URAT1), responsible for reabsorbing over 90% of uric acid in the kidneys and thereby preventing its accumulation in the blood—which can lead to gout—is a crucial target for the development of new anti-hyperuricemic medications. Currently, lesinurad are the only URAT1 inhibitor approved by the Food and Drug Administration, highlighting the need for additional treatments. Machine learning (ML) can enhance structure-based virtual screening for discovering novel URAT1 inhibitors by predicting protein-ligand interactions. This study provides quantitative assessment of the prediction performance of the latest ML models for URAT1 using a unique dataset of 3D URAT1-ligand structures generated by AlphaFold2 and Smina, a fork of AutoDock Vina v1.1.2, as input data and pIC50 as label data. The dataset includes high-active URAT1 inhibitors, low-active URAT1 inhibitors, and simulated inactive structures generated by DeepCoy (Imrie et al., 2021) with properties matching those of the high-active inhibitors. The ratio of active to inactive compounds is maintained at 1:100. The systematic evaluations with respect to AUC, Enrichment Factor and Normalized Enrichment Factor are reported for the ML methods, such as supervised ML with PLEC fingerprints (Wójcikowskiet al, 2019), convolutional neural networks (McNutt et al, 2021), and geometric interaction graph neural network (GIGN) (Yang et al, 2023).