P07-16
Development of a compound pre-screening method based on docking of fragments
Masayoshi SHIMIZU *, Keisuke YANAGISAWA, Yutaka AKIYAMA
School of Computing, Institute of Science Tokyo
( * E-mail: shimizu@bi.c.titech.ac.jp )
Structure-based virtual screening (SBVS) is a computational technique to select compounds using 3D structural information of target proteins and compounds. Recently, more than several hundred million compounds are registered in compound database [1], and thus, a method to select from a large number of compounds at high speed is required. Protein-ligand docking is often used in SBVS, but it has high computational complexity and development of faster methods is highly demanded. One of the ways to realize faster calculation is to focus on the fragments of compounds. Since, for example, 28 million compounds can be represented by only 263 thousand fragments [2], fragment-based ligand docking approach enables evaluation of compounds with a fragment set that has fewer number of types than that of the compound set. However, fragment-based docking still spends several CPU core years to evaluate hundreds of millions of compounds.
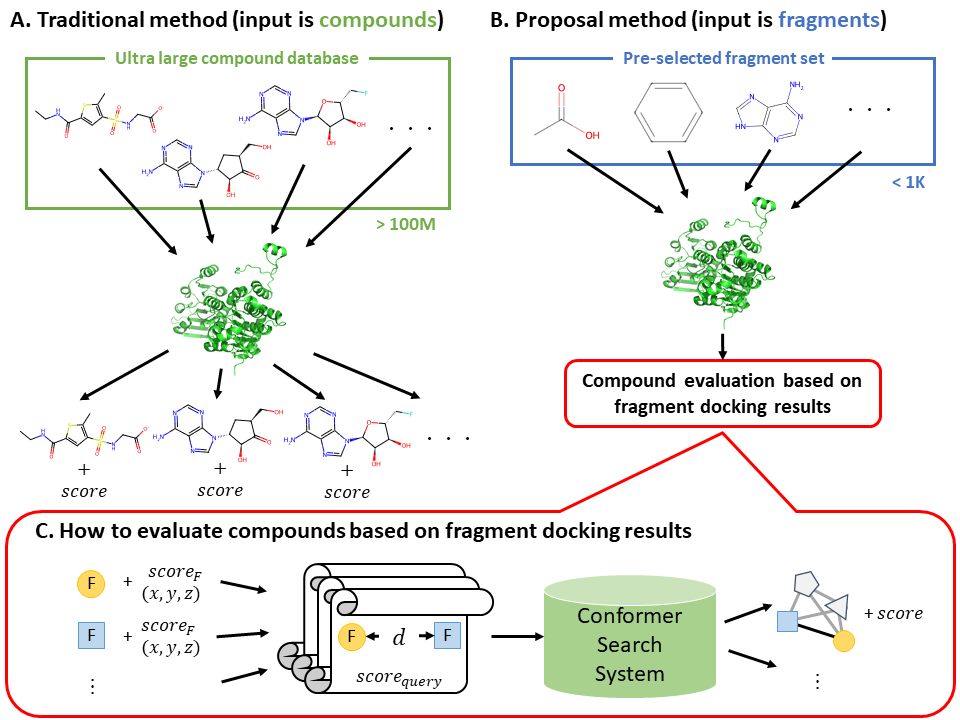
In this study, unlike conventional fragment-based methods that the compounds are used as input (Figure A), a pre-selected representative fragment set is used as input (Figure B). We proposed a compound pre-screening method that searches for possible conformations of known compounds based on the relative distance between spatial arrangement of pairs of representative fragments and selects those compounds as candidates (Figure C).
In computational experiments with six targets in DUD-E, we confirmed that pre-screening was up to 60 times faster than existing protein-ligand docking tools like REstretto [3]. On the other hand, the prediction accuracy still needs to be improved. A possible approach to improve accuracy might be to consider arrangements of more than three fragments.
[1] BI Tingle, et al., JCIM 63, 1166-1176, 2023.
[2] K Yanagisawa, et al., Bioinformatics 33, 3836-3843, 2017.
[3] K Yanagisawa, et al., ACS omega 7, 30265-30274, 2022.