P07-14
QUBO Problem Formulation of Fragment-Based Protein–Compound Flexible Docking
Keisuke YANAGISAWA *1, 2, Takuya FUJIE3, Kazuki TAKABATAKE4, Yutaka AKIYAMA3
1Department of Computer Science, School of Computing, Institute of Science Tokyo
2Middle Molecule IT-Based Drug Discovery Laboratory (MIDL), Institute of Science Tokyo
3Ahead Biocomputing, Co., Ltd.
4Toshiba Digital Solutions Corporation
( * E-mail: yanagisawa@c.titech.ac.jp )
Protein–ligand docking plays a significant role in structure-based virtual screening. In recent years, the size of compound libraries has been growing explosively, resulting in a demand of faster docking methods. One of the promising approaches is quantum annealing, which has been attracting attention. Quantum annealing efficiently solves combinatorial optimization problems and is suitable for docking because docking is an optimization problem searching for the best scoring pose. In quantum annealing, it is important to formulate this docking as a quadratic unconstrained binary optimization (QUBO) problem to obtain solutions, and thus, such formulations have been attempted. However, most previous studies did not consider the internal degrees of freedom of the compound that is mandatory and essential.
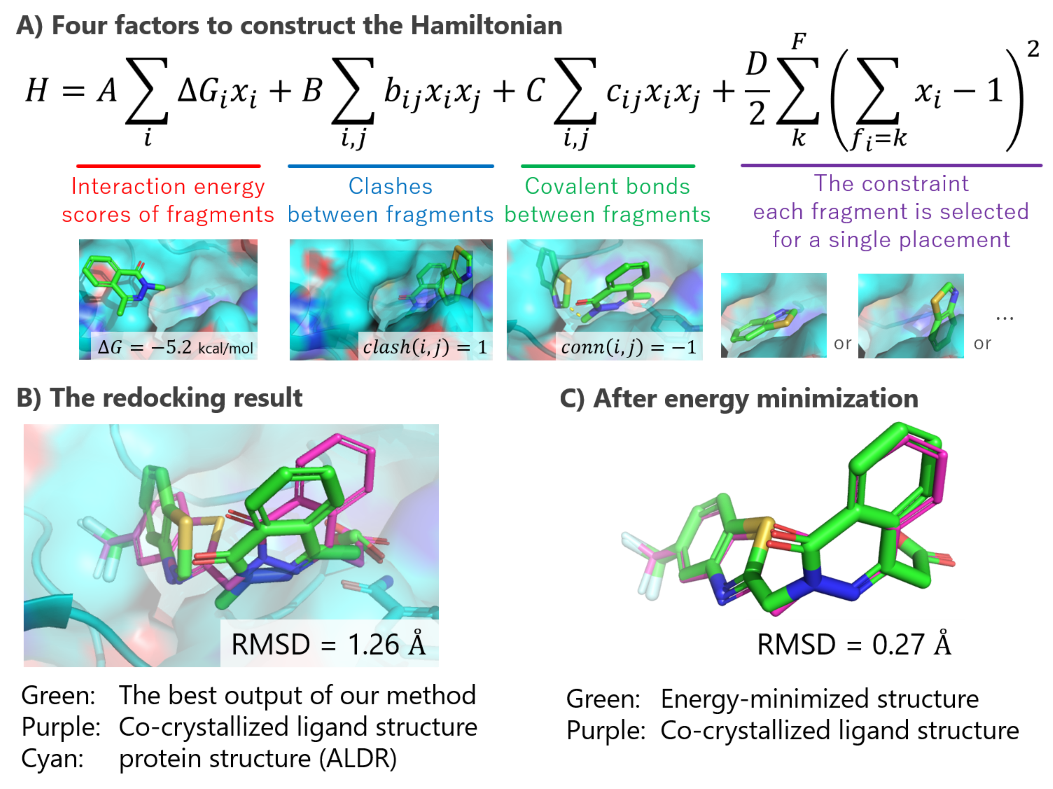
In this study, we formulated fragment-based protein–ligand flexible docking, considering the internal degrees of freedom of the compound by focusing on fragments (rigid chemical substructures of compounds) as a QUBO problem. [1] We introduced four factors essential for fragment–based docking in the Hamiltonian: (1) interaction energy between the target protein and each fragment, (2) clashes between fragments, (3) covalent bonds between fragments, and (4) the constraint that each fragment of the compound is selected for a single placement (Figure A).
We also implemented a proof-of-concept system and conducted redocking for the protein–compound complex structure of Aldose reductase using SQBM+, which is a simulated quantum annealer. The predicted binding pose reconstructed from the best solution was near-native (Figure B, RMSD = 1.26 Å), which can be further improved (Figure C, RMSD = 0.27 Å) using conventional energy minimization. The results indicate the validity of our QUBO problem formulation.
[1] K Yanagisawa, T Fujie, K Takabatake, Y Akiyama. QUBO Problem Formulation of Fragment-Based Protein-Ligand Flexible Docking. Entropy 26, 397, 2024. DOI: 10.3390/e26050397