P07-10
Virtual validation and the efficient learning methods exploration in federated learning (FL) for drug development research
Ziwei ZHOU *1, Reiko WATANABE1, Masataka KURODA2, 3, Kenji MIZUGUCHI1
1 Institute for Protein Research, Osaka University
2National Institutes of Biomedical Innovation, Health and Nutrition, 3Mitsubishi Tanabe Pharma Corporation
( * E-mail: u631859f@ecs.osaka-u.ac.jp )
Applying Machine Learning (ML) methods in drug development requires large-scale and high-quality experimental data, which is hard to obtain from public sources. It is expected that collaboration with multiple pharmaceutical companies will significantly facilitate data collection. However, sharing data with another research group needs to involve the risk of data leaking and even causing economic losses. Intellectual property issues are a major barrier to business collaborations and public-private partnerships. Federated Learning (FL) is an ML method that allows multi-source data join training via the client’s server, which solves intellectual property problems to a certain extent. There is a possibility that differences in chemical space, number of data sets, and data heterogeneity among each client may worsen the consistency and convergence of the training model. Although it is essential to consider appropriate learning conditions in FL, the impact of data differences among clients on FL has not yet been fully explored.
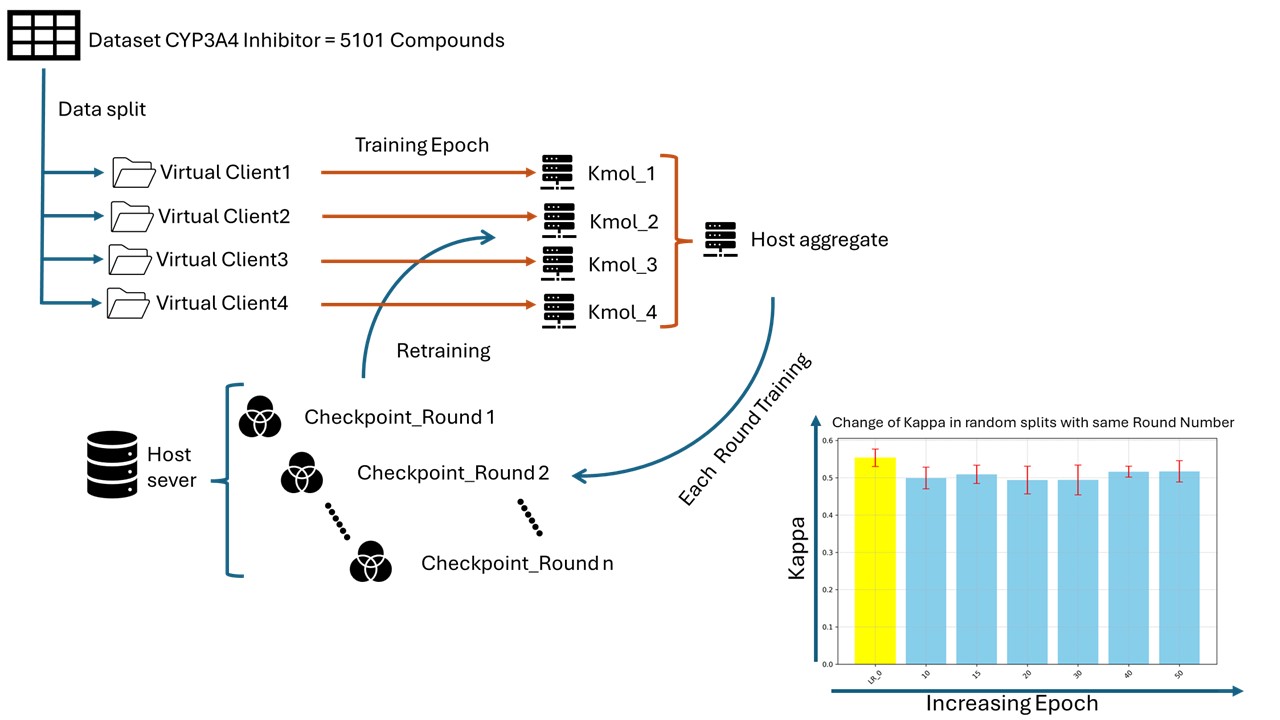
This research aims to validate the feasibility of FL and identify efficient learning methods. The study is based on CYP3A4 inhibition data with a 10µM threshold containing 5101 compounds. The datasets were split using three different segmentations: random splits and splits showing different chemical spaces with and without unifying the number of data sets to simulate real situations. Then classification models for CYP3A4 inhibition were constructed using graph convolutional networks at virtual FL in different training conditions such as hyperparameter setting in epoch, round numbers and other settings.
It showed that hyperparameters will largely influence the performance of FL. In the best hyperparameters setting used in full dataset training, FL models with random split could reach the equivalent performance of local training. We will discuss how we can optimize the learning condition in FL
Our finding would make it possible to present more suitable learning conditions for future FL implementation and provide a possible method to boost model training speed via FL methods.