P07-09
Development of a Massive Fluorogenic Probe Library Based on Bayesian Optimization toward the Discovery of Novel Biomarker Enzymes
Daiki ISHIMOTO *1, Ryo TACHIBANA1, Yasuteru URANO1, 2
1Laboratory of Chemistry and Biology, Graduate School of Pharmaceutical Sciences, The University of Tokyo
2Department of Chemical Biology and Molecular Imaging, Biomedical Engineering, Radiology and Biomedical Engineering, Graduate School of Medicine, The University of Tokyo
( * E-mail: d-ishimoto2001@g.ecc.u-tokyo.ac.jp )
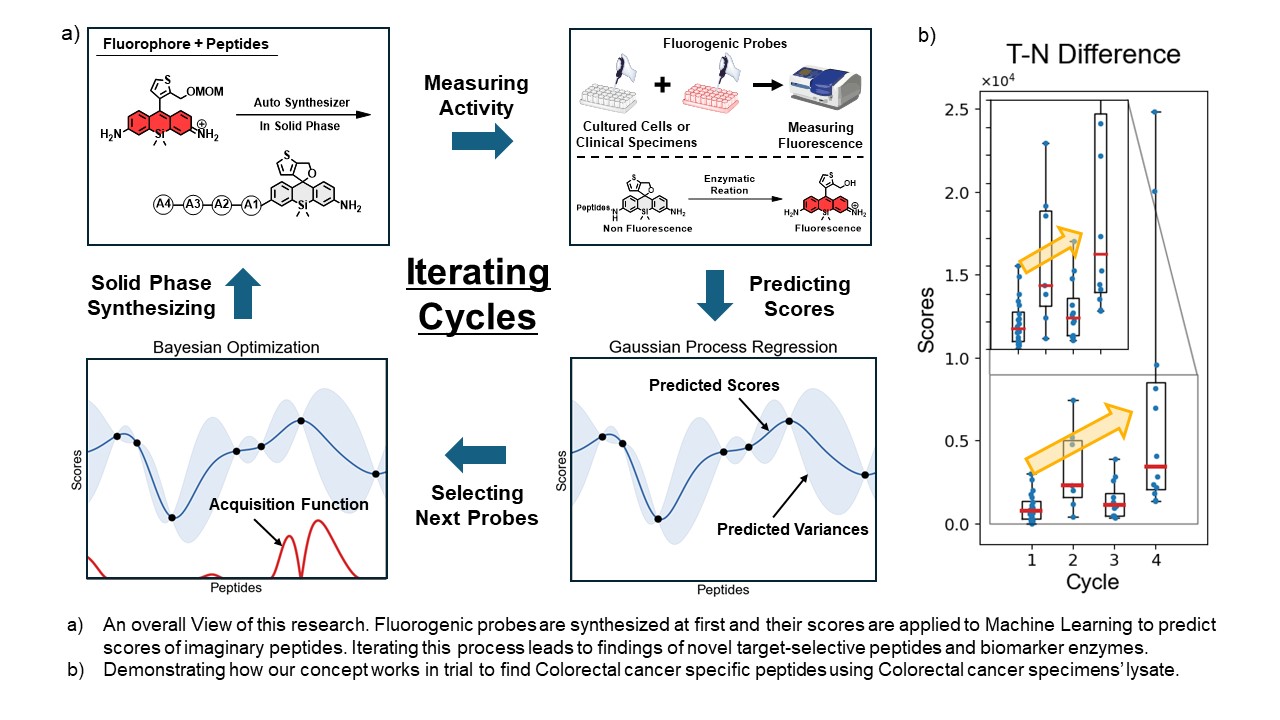
Drastic treatment for many types of cancer is to resect whole tumor regions. However, it is difficult to distinguish between tumor and non-tumor tissues with unaided human eyes and cancer tissues left in patients’ body cause recurrence and metastasis which lead to poor prognosis. For these reasons, clinical needs for cancer-selective visualizing tools are growing and among those are fluorogenic probes. These fluorogenic probes are non-fluorescence and enzymatic reactions at target sites turn fluorescence on. In previous research, a fluorogenic probe library was constructed to find novel tumor-selective biomarker enzymes and as a result, promising probes and biomarker enzymes were found for tumor-selective visualization.
However, present ways of developing target-specific probes have many problems. For example, since we must synthesize all candidate probes, varieties of target enzymes and probes’ structure are limited. In addition to this practical problem, tumor-selective enzymes need to be known well in advance to find its selective probes. Considering how many enzymes and its substrates are, establishing conclusive methods for searching target-specific enzyme-substrate pairs would be difficult. Here, we propose one solution to this challenge by using Machine Learning techniques: Gaussian Process(GPR) and Bayesian Optimization(BO).
BO leverages results of GPR, which is used for predicting scores of imaginary probes. In BO, Acquisition Function plays a key role in deciding which probes are more likely to show better scores among imaginary probes. There are many Acquisition Functions like Probability Improvement, Expected Improvement and Predicted Entropy Search as major examples. They commonly use predicted scores to estimate to what extent each imaginary probe improves scores and regression accuracy. Next, we synthesize suggested probes by BO and evaluate its scores in experiments. After that, BO are applied again to recalculate imaginary probes’ improvements. By iterating this process, predicting and evaluating, we could reach probes that have desired properties without evaluating a numerous amount of candidate probes.
In this research, we employed a fluorophore with bright fluorescence in the visible wavelength region: HMRR (hydroxymethyl rhodamine red). HMRR with tetra peptides consist of 20 essential amino acids are adapted as candidate probes which have 160,000 (204) kinds in total. It is totally impossible to synthesize and evaluate all these probes in real and these high-dimensional probes have yet to be explored well. Making use of Machine Learning techniques introduced above, finding novel biomarker-probe pairs becomes easier from a great variety of candidates with a minimum number of real experiments. We hope our research contributes to explain how Machine Learning system works in Chemical Biology.