P07-08
A Dirichlet diffusion model for generation of high-quality antimicrobial peptide sequences
Koichi OKI *1, Shuto HAYASHI2, Jun KOSEKI3, Teppei SHIMAMURA1, 2
1Graduate School of Medicine, Division of Systems Biology, Nagoya University
2Medical Research Institute, Institute of Science Tokyo
3Cellular and Molecular Biotechnology Research Institute, National Institute of Advanced Industrial Science and Technology (AIST)
( * E-mail: oki.koichi.c5@s.mail.nagoya-u.ac.jp )
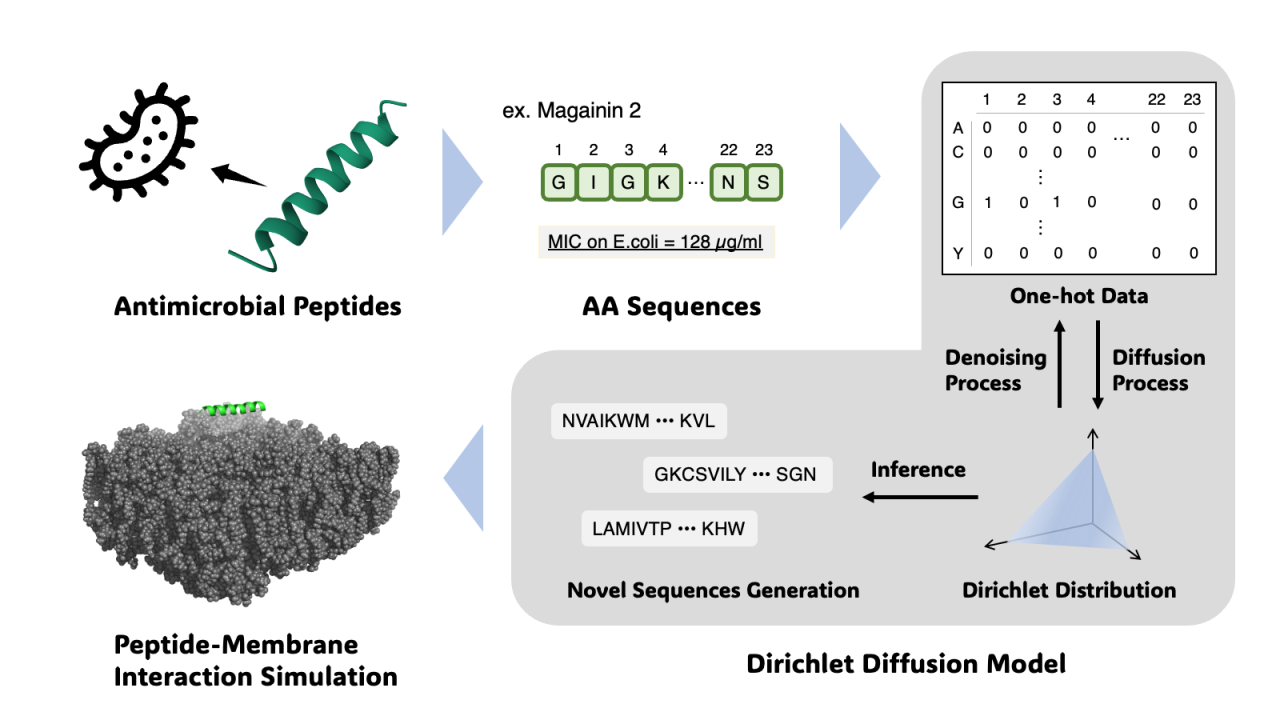
The misuse of antibiotics has led to the rise of drug-resistant bacteria, projected to become the leading cause of death globally by 2050. Antimicrobial peptides (AMPs), which function differently from traditional small-molecule drugs, have gained attention due to their ability to delay bacterial resistance. AMPs interact with bacterial membranes due to their amphipathic nature, causing cell lysis. While machine learning has been explored for generating novel peptides, the discrete nature of peptide sequences makes feature extraction and quality generation challenging. Current AMP generative models, including autoencoders, diffusion models, and transformers, tackle this issue by mimicking original sequence characteristics. However, these models often label peptides in datasets using a fixed threshold for Minimum Inhibitory Concentration (MIC), leading to low-resolution MIC data. For more effective peptide generation, MIC should be treated as continuous values.
We developed a deep learning-based method for generating high-quality AMP sequences using a Dirichlet diffusion score model, known for its strong performance in discrete data generation. Our model incorporates continuous MIC values during training and sequence generation, establishing a framework that uses molecular dynamics (MD) simulations to verify AMP and bacterial membrane interactions.
The model was trained on sequences with confirmed antimicrobial activity from the DBAASP database. The generated AMPs retained key characteristics of the original sequences, such as amino acid composition and physicochemical property distributions. By using continuous MIC values and guiding the model to generate sequences with lower MIC values, our method efficiently produces highly active peptides compared to existing models.
We also conducted MD simulations on the generated AMP candidates. Although AMPs primarily exert their effects through interactions with bacterial membranes, simulating a cell’s lipid bilayer is computationally expensive. We combined coarse-grained and all-atom simulations, enabling high-throughput and effective evaluation of many generated peptides.
This study marks the first application of the Dirichlet diffusion score model in AMP sequence generation and shows that integrating continuous MIC values with multiscale simulation techniques can enhance peptide design significantly. Further validation through experiments with actual bacteria or animals, including assessments of toxicity and hemolytic effects, is necessary to confirm the quality of the generated AMPs.