P07-06
De novo PROTAC linker design to enhance cell membrane permeability based on a data-driven method
Yuki MURAKAMI *1, Shoichi ISHIDA1, Yosuke DEMIZU1, 2, Kei TERAYAMA1
1Graduate School of Medical Life Science, Yokohama City University
2Division of Organic Chemistry, National Institute of Health Science
( * E-mail: w245513c@yokohama-cu.ac.jp )
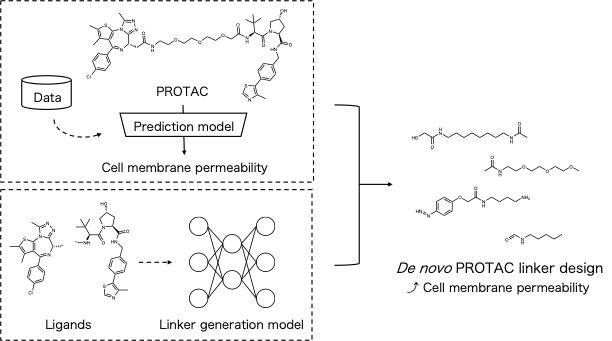
Proteolysis targeting chimeras (PROTACs) have garnered significant interest as next-generation therapeutics capable of degrading disease-related proteins of interest (POIs) [1]. PROTACs are chimeric molecules consisting of an E3 ligase-binding moiety, a POI ligand, and a linker. The linker structures of PROTACs significantly influence biodegradation efficiency and physicochemical properties including cell membrane permeability [2]. Optimizing the linker structures is crucial for improving both biodegradation efficiency and physicochemical properties. Recently, many machine learning-based methods that generate PROTAC linkers with various improved properties, such as linker length, logP, and three-dimensional binding conformations, have been developed [2]. However, a PROTAC linker design method to improve cell membrane permeability, which is one of limitations of PROTACs [3], remains undeveloped.
Here, we developed a machine learning-based PROTAC linker design method to improve cell membrane permeability. To evaluate the cell membrane permeability of PROTACs with the designed linker, we constructed a prediction model using a machine learning approach based on public experimental data. We designed PROTAC linkers by combining molecular generative models [4,5], and the prediction model to improve cell membrane permeability. At this conference, we report both the proposed method and the results.
[1] Tsai Jonathan M.; et al., Targeted protein degradation: from mechanisms to clinic, Nature Reviews Molecular Cell Biology, 2024, 1-18.
[2] Dong Yawen; et al., Characteristic roadmap of linker governs the rational design of PROTACs, Acta Pharmaceutica Sinica B, 2024.
[3] Apprato Giulia; et al., Exploring the chemical space of orally bioavailable PROTACs, Drug Discovery Today, 2024, 103917.
[4] Ishida Shoichi; et al., ChemTSv2: Functional molecular design using de novo molecule generator, Wiley Interdisciplinary Reviews: Computational Molecular Science, 2023, 13, e1680.
[5] Zheng Shuangjia; et al., Accelerated rational PROTAC design via deep learning and molecular simulations, Nature Machine Intelligence, 2022, 4, 739-748.