P07-03
Open Source Program Github and Its Application in Drug Discovery
Kiyoshi HASEGAWA *, Yuya SEKI, Yu LIU, Yukiyo ITO
Division of Informatics Promotion, TECHNOPRO R&D company
( * E-mail: Hasegawa.Kiyoshi@technopro.com )
GitHub is an open-source program aimed at validating the paper and further improving its algorithm when researchers have submitted to journal. It was recently recognized that Github programs are useful tools in drug discovery toward pre-clininal phase. For example, in cheminformatics, this includes generating compound structures within proteins and predicting the activity and physical properties of compounds with chemical interpretations. By combining these two approaches, it is possible to obtain new chemical structures with improved compound profiles. In bioinformatics, this includes single-cell analysis and cell differentiation analysis with time series. Additionally, it encompasses the generation of sequences for active antibodies and peptides.
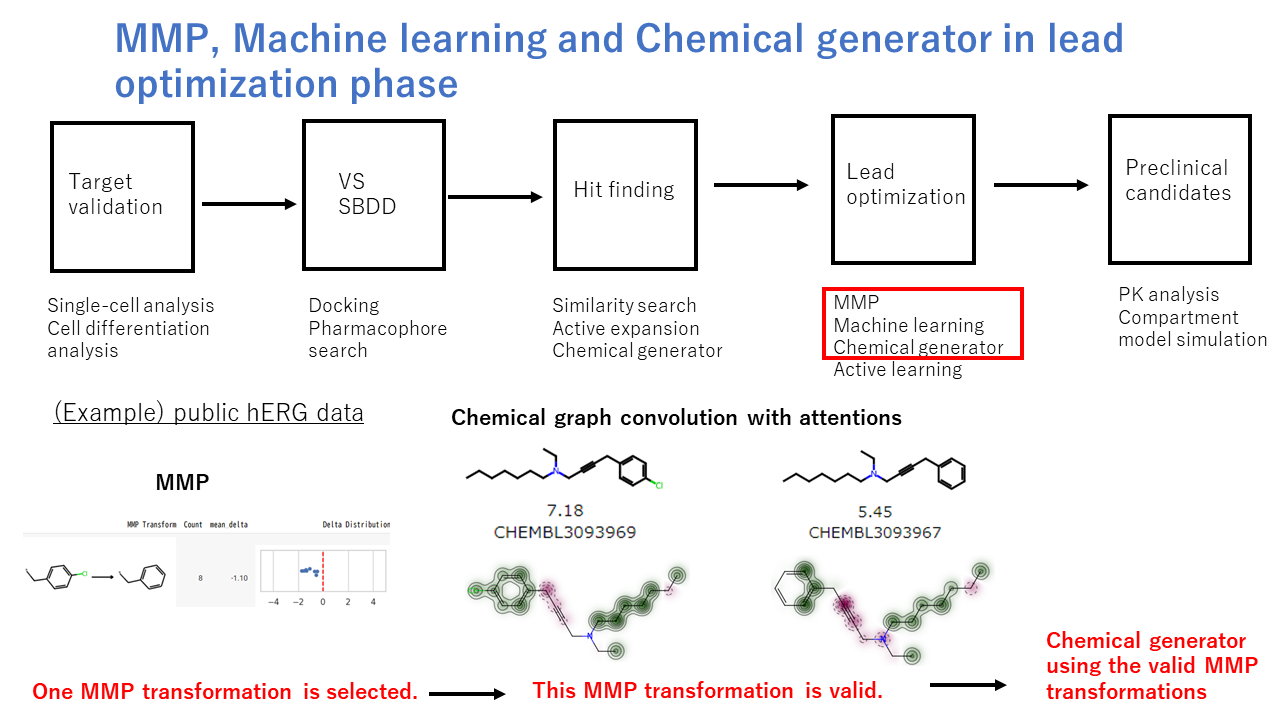
We focus on the optimization phase to customize MMP (matched molecular pairs), machine learning and chemical generator tools in cheminformatics field. First of all, all possible MMP transformation rules are extracted from RDKit and Pandas libraries. Then, each MMP transformation is validated on two aspects. One is how to fit the prediction values from chemical graph convolution model to the observed values. DeepChem library is used for building chemical graph convolution model. Second is whether the shaded colors derived the attention values on chemical structure is matched to SAR (structure-activity relationships) and chemist’s intuitions. The attention values are calculated from the graph convolution weights and the contributions of chemical fragments to activities. Third, if above two criteria would be passed, the remaining MMP transformations are processed to chemical generator framework. Keeping the core structure, MMP transformations are used to generate possible chemical libraries.
This optimization strategy will be applied to other ADME properties other than hERG inhibition data. Also, this strategy is further extended to multi-optimization solutions when the MMP transformations from possible ADME properties would be prepared in advance.