P07-02
Discovery of a new histone deacetylase 8 inhibitor using machine learning-aided drug screening
Yasunobu YAMASHITA *, Atika NURANI, Yuuki TAKI, Yuri TAKADA, Yukihiro ITOH, Takayoshi SUZUKI
SANKEN, Osaka University
( * E-mail: yyamashita@sanken.osaka-u.ac.jp )
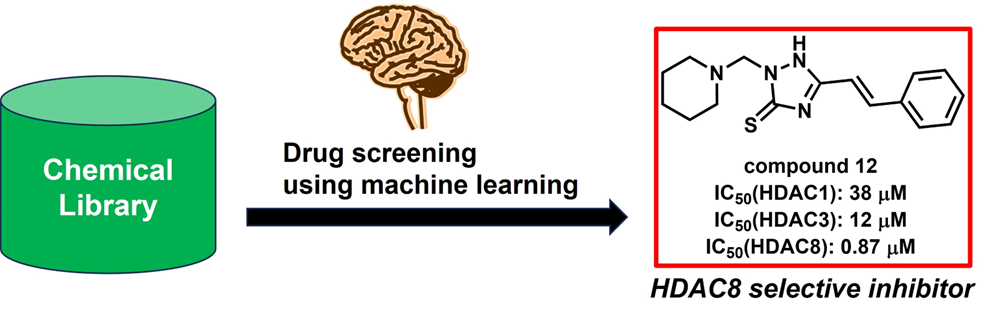
Histone deacetylase 8 (HDAC8) is a zinc-dependent enzyme that catalyzes the deacetylation of non-histone proteins and is involved in cancer development. HDAC8 inhibitors are promising candidates as anticancer agents. However, most reported HDAC8 inhibitors contain a hydroxamic acid moiety, which is often associated with mutagenicity. Therefore, we used machine learning for drug screening to identify non-hydroxamic acid HDAC8 inhibitors. In this study, we established a prediction model based on the random forest (RF) algorithm for screening HDAC8 inhibitors, as it exhibited the best predictive accuracy on a training dataset augmented with data generated by the synthetic minority over-sampling technique (SMOTE). Using the trained RF-SMOTE model, we screened the Osaka University library for compounds and selected 50 virtual hits. However, none of these initial 50 hits showed HDAC8-inhibitory activity. In a second screening, utilizing an RF-SMOTE model retrained with a dataset including these 50 inactive compounds, we identified non-hydroxamic acid compound 12 as an HDAC8 inhibitor with an IC50 of 0.87 μM.1) Interestingly, its IC50 values for HDAC1 and HDAC3 inhibitory activity were 38 μM and 12 μM, respectively, demonstrating that compound 12 has high selectivity for HDAC8. Through the use of machine learning, we expanded the chemical space for HDAC8 inhibitors and identified non-hydroxamic acid 12 as a novel HDAC8 selective inhibitor.
Reference
1) A. Nurani, Y. Yamashita, Y. Taki, Y. Takada, Y. Itoh, T. Suzuki. Chem. Pharm. Bull. 2024, 72, 173–178.