P06-11
Elucidation of Stabilization Mechanisms of Intrabodies Based on Statistical Thermodynamics
Koki HATTORI *1, Yuto HOSHI1, Hiroshi YUKAWA1, 2, Satoshi OGASAWARA1, Masahiro KINOSHITA1, 3, Takeshi MURATA1, Satoshi YASUDA1
1Graduate School of Science and Engineering, Chiba University
2National Institutes for Quantum Science and Technology
3Institute of Advanced Energy, Kyoto University
( * E-mail: 23wm2615@student.gs.chiba-u.jp )
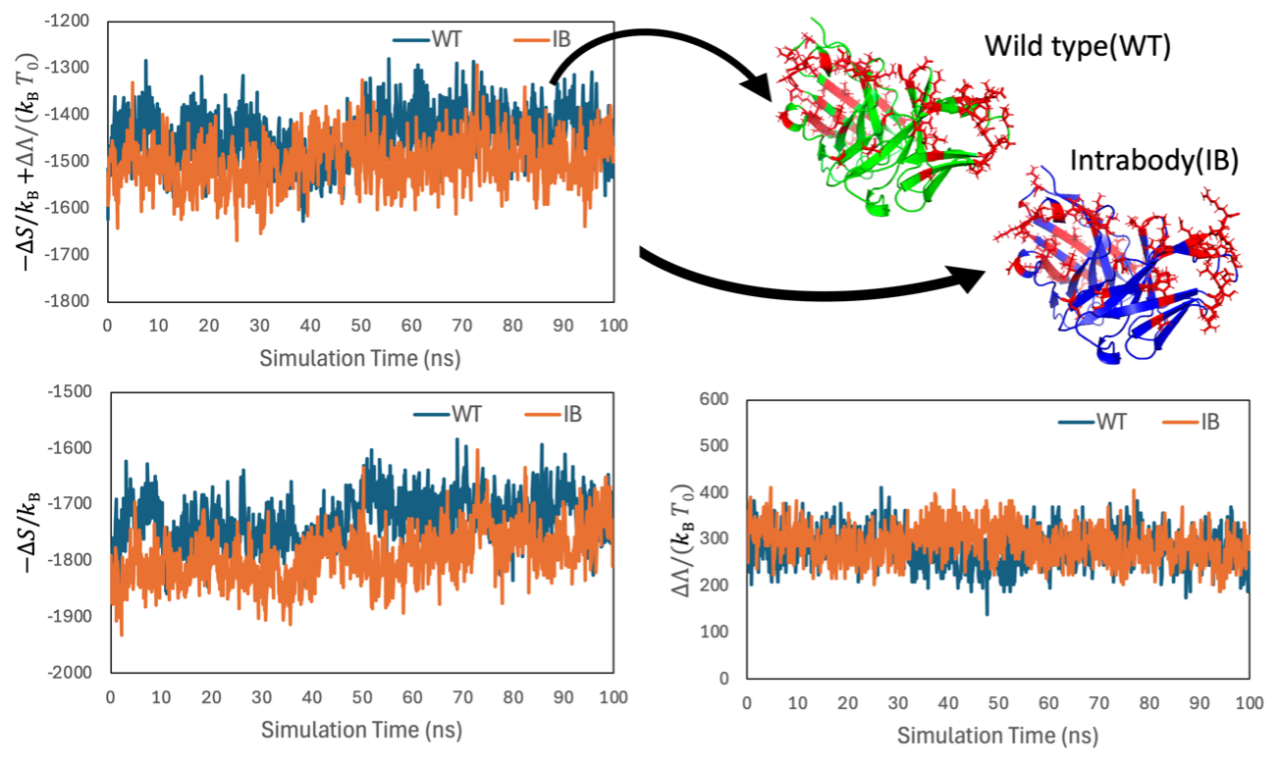
Single-chain variable fragments (scFvs), which are recombinant antibody fragments consisting of only the variable light chain and variable heavy chain domains covalently connected to one another by a short polypeptide linker, are expected to be used in pharmaceuticals and other applications. Recently, stabilized multimutant scFvs, known as intrabodies, constructed using knowledge-based methods have been reported, but the details of their stabilization mechanisms have remained unclear. In our earlier study, Kinoshita et al. developed the free energy function (FEF) that can accurately evaluate the thermal stability of proteins. Our FEF consists of the entropy term S and the energy term Λ. S represents the water entropy change arising from the excluded volume effect and is calculated using an original method based on statistical thermodynamic theory. Λ, on the other hand, represents the energy change due to dehydration and is calculated by counting hydrogen bonds within proteins and between water and proteins. In this study, we have elucidated the stabilization mechanism of intrabodies by calculating the free energy changes upon folding using FEF for four intrabodies and their wild-type structures. The structure models of the intrabodies and the wild type were constructed using AlphaFold2. Fluctuations of the antibody were taken into account using molecular dynamics simulations. The results of this study are as follows: Intrabodies have a larger water entropy gain upon folding than the wild types. This indicates that closer packing of side chains is achieved by the amino acid mutations of intrabodies. We have also identified which amino acid mutations among the multimutants play a particularly important role in stabilization. Based on these findings, we are now investigating further stabilizing mutants of intrabodies.