P06-09
Investigation of the trends and the potential in drug development for rare and intractable diseases based on the KEGG NETWORK
Mao TANABE *, Makoto HIRATA, Ryuichi SAKATE
Laboratory of Rare Disease Information and Resource Library, Center for Intractable Diseases and ImmunoGenomics (CiDIG), National Institutes of Biomedical Innovation, Health and Nutrition
( * E-mail: mtanabe@nibiohn.go.jp )
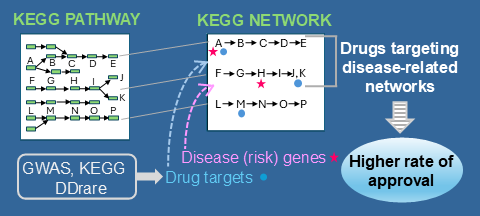
For many rare and intractable diseases (RIDs), the pathophysiological mechanisms still remain unexplained and there are few drugs for the treatment of these diseases. An understanding of approved drugs is important to improve drug development. In DDrare (Database of Drug Development for Rare Diseases) [1], the targets of drugs in clinical trials are mapped to the KEGG PATHWAY to be grasped on molecular networks. In this study, to understand the relationship between drug targets and disease genes more precisely, we mapped them to the KEGG NETWORK (networks) defined as functionally meaningful segments of pathways [2,3]. We found that disease genes tended to be included in networks characteristic for each disease group, whereas drug targets were mapped to networks common to many disease groups. The number of drugs targeting the networks containing disease genes was small in every disease group. However, because several studies have recently addressed that the drugs that target proteins with direct genetic evidence of disease association and their molecular partners are more likely to be approved [4,5], we confirmed the results using the KEGG NETWORK and integrating the risk genes obtained from the latest GWAS data. The results were clearer and more detailed than those of previous studies. Considering the findings in literature, the fact that a drug targeting a network with disease genes has been approved suggests the perturbation of the network by specific causes, but not always by the disease genes in the network. The knowledge acquired in this study shows the possibility that the perturbed network, and in turn precise targets of drug for a RID can be found by obtaining GWAS data, other omics data, or the information about environment factors of the disease, and by mapping them to the functionally meaningful segments of pathways such as the KEGG NETWORK.
[1] DDrare: Database of Drug Development for Rare Diseases, https://ddrare.nibiohn.go.jp/
[2] KEGG: Kyoto Encyclopedia of Genes and Genomes, https://www.kegg.jp/kegg/
[3] Tanabe M., Hirata M., and Sakate R., Trends in drug development for rare and intractable diseases based on the KEGG NETWORK. NAR Molecular Medicine. 2024; 1
[4] Nelson M.R., Tipney H., Painter J.L., et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015; 47:856-60.
[5] Okada Y., Wu D., Trynka G., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014; 506:376-81.