P06-04
Impact of Intramolecular Hydrogen Bonds on Permeability Glycoprotein Mediated Transportation
Yulong GOU *1, Suyong RE2, Kenji MIZUGUCHI1, Chioko NAGAO1
1Laboratory for Computational Biology , Osaka University, Insitute for Protein Research
2National Institute of Biomedical Innovation, Health and Nutrition
( * E-mail: gouyl96@protein.osaka-u.ac.jp )
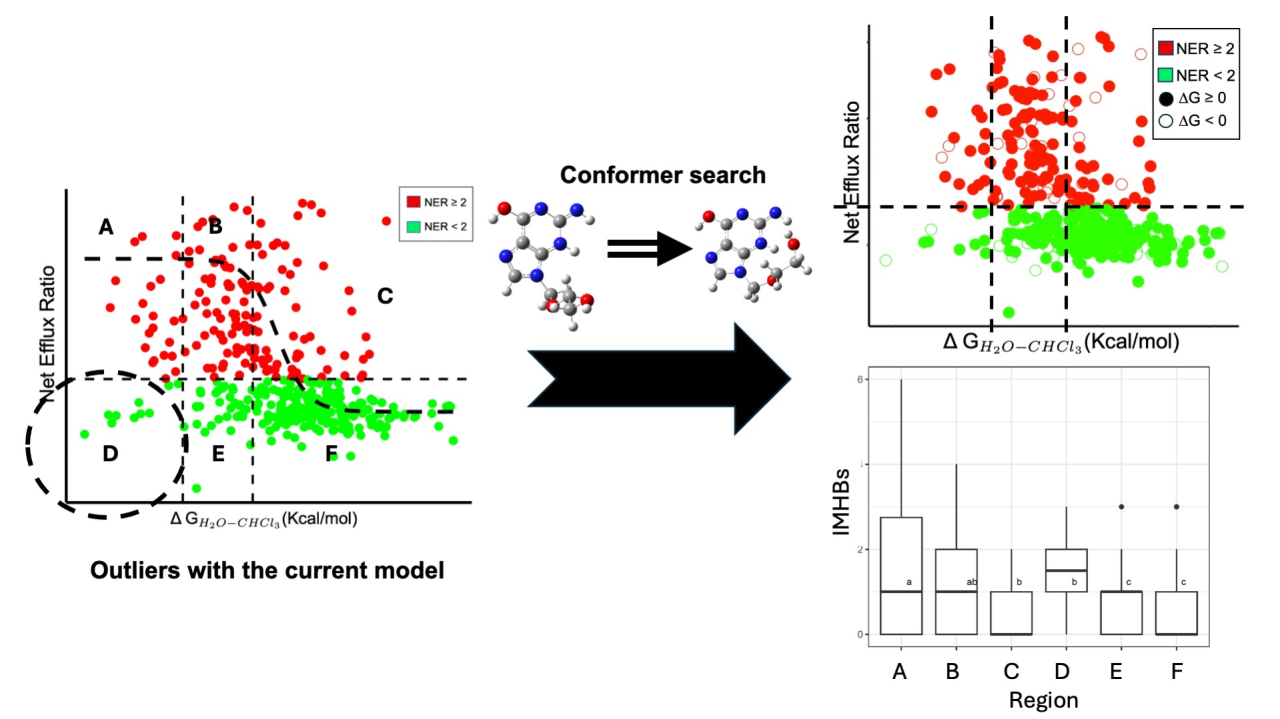
Permeability glycoprotein (P-gp) is widely expressed in many cells related to pharmacokinetics and physical barriers in human body. P-gp pumps diverse substances and toxins out of cells through ATP-mediated conformational changes. Overexpression of P-gp lead to multidrug resistance. The efflux ratio (ER) is often used to determine the ability of P-gp-mediated efflux in drug discovery. Due to the lack of large-scale data, a de novo computational method based on solvation free energy was developed1, which is useful for predicting whether a drug compound is a P-gp substrate or not. In this method, the net ER is related to the solvation free energy by a sigmoidal function. Recently, we evaluated this method, using in-house data set of 397 compounds, and found that there are non-negligible outliers, although this method provides a good result on several compounds2. The outliers typically involve the compounds with the large solvation free energy, the low efflux ratio, and the high potential to donate hydrogen bond. Since the intermolecular hydrogen bonds (IMHBs) can affect the solvation free energy, we re-evaluated the method by rigorously considering the IMHBs. We employed the RDKit3 to generated 100 conformers for each of 397 compounds and obtained the optimal structures. Many of the obtained structures contain IMHB regardless of substrates or non-substrates. Notably, the outliers have more IMHB than other compounds. We found that accounting for IMHBs in the calculation of solvation free energies decrease the values compared to the original evaluation without the conformer search, which slightly reduces the number of outliers. Considering the non-negligible outliers, explicitly accounting for the specific interactions between the compounds and P-gp could further improve the prediction model.
1. Gunaydin H. et al. ACS Med.l Chem. Lett. 2013, 4 (1), 108-112.
2. Gou Y. et al. ACS Med.l Chem. Lett. 2023, 15 (1), 54-59.
3. RDKit: Open-source cheminformatics; http://www.rdkit.org