P05-05
Addressing Common Metabolism Problems in Drug Discovery with in Silico Methods
Sumie TAJIMA *1, Daniel A. BARR2, Shiori TAKASE1, Mario ÖEREN2, Peter A. HUNT2, Tomáš CHRIEN2, Tamsin E. MANSLEY2, Matthew D. SEGALL2
1HULINKS Inc.
2Optibrium Limited
( * E-mail: tajima@hulinks.co.jp )
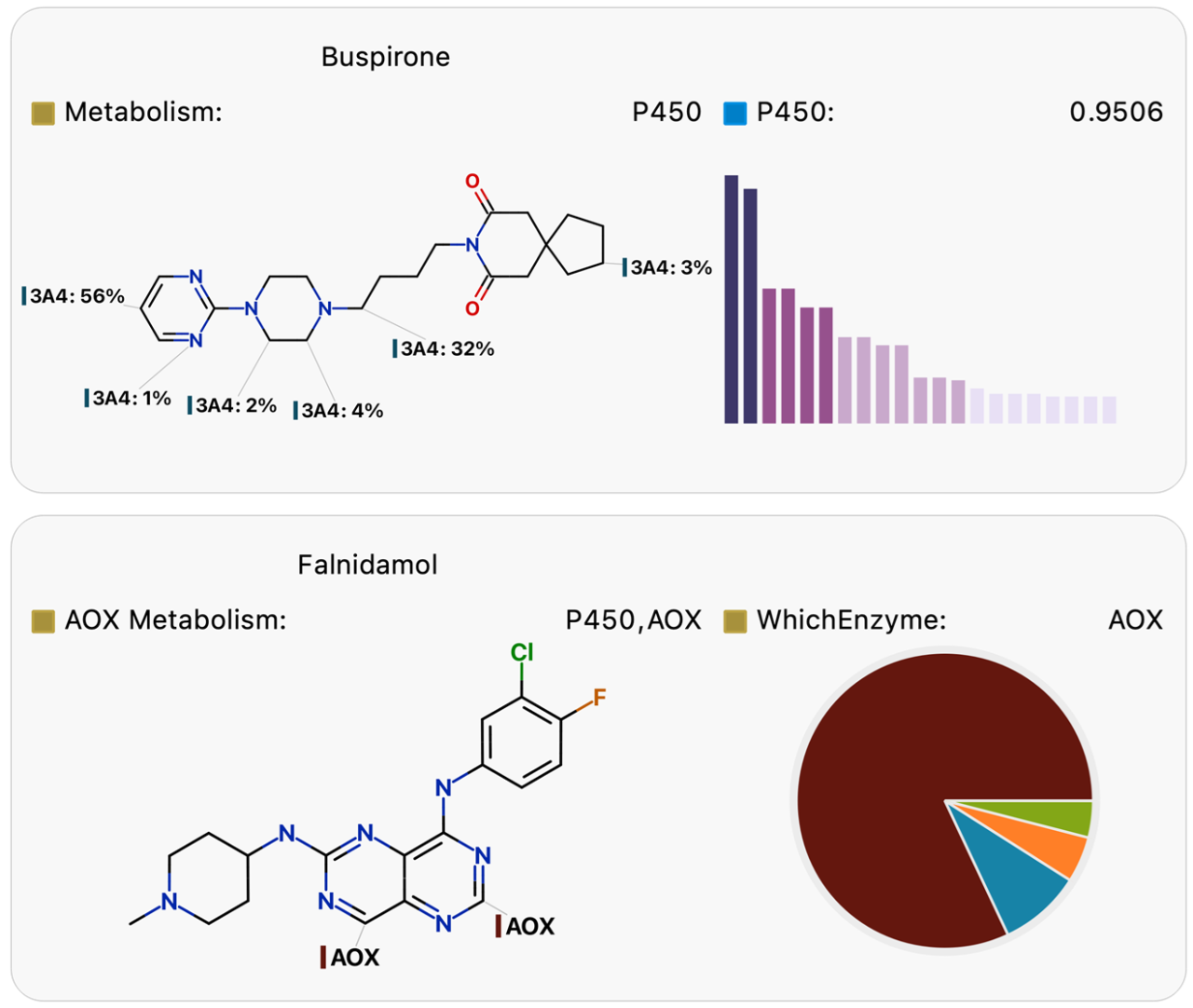
In silico metabolism prediction can address critical questions to guide lead optimization. Using several case studies, we demonstrate the application of these models to address design challenges involving metabolic (in)stability, the formation of reactive and/or toxic intermediates, and to mitigate the risk of genetic polymorphisms and drug-drug interactions. In addition, we illustrate how these models can inform the selection of in vitro and in vivo pre-clinical experiments to avoid surprises in late-stage trials. Furthermore, accurate predictions of metabolite profiles early in the discovery process provide essential guidance for drug design. Optibrium’s mechanistic metabolism models cover metabolism by P450, AOX, FMO, UGT, and SULT enzymes1-5. By combining these models, metabolic pathway analysis proposes the most likely metabolites with greater precision than other methods, assisting in metabolite identification studies and enabling potentially active, reactive, or toxic metabolites to be identified6.
References
1) Mario Öeren, Peter J. Walton, James Suri, David J. Ponting, Peter A. Hunt and Matthew D. Segall, (2022) J. Med. Chem. 65(20) pp. 1406-1408
2) Mario Öeren, Sylvia C. Kaempf, David J. Ponting, Peter A. Hunt and Matthew D. Segall, (2023) J. Chem. Inf. Model. 63(11) pp. 3340-3349
3) Mario Öeren, Peter J. Walton, Peter A. Hunt, David J. Ponting and Matthew D. Segall, (2021) J. Comput.-Aided Mol. Des. 35(4) pp. 541-555
4) Jonathan D. Tyzack, Peter A. Hunt and Matthew D. Segall, (2016) J. Chem. Inf. Model. 56(1) pp. 2180-2193
5) Peter A. Hunt, Matthew D. Segall & Jonathan D. Tyzack, (2018) J. Comput.-Aided Mol. Des., 32 pp. 537-546
6) Mario Öeren, Peter A. Hunt, Charlotte E. Wharrick, Hamed Tabatabaei Ghomi and Matthew D. Segall, (2023) Xenobiotica DOI: 10.1080/00498254.2023.2284251