P04-03
Development of the Cryptic Site searching method with Mixed-solvent molecular dynamics and Topological data analyses methods
Jun KOSEKI *1, Motono CHIE1, 2, Yanagiswa KEISUKE3, 4, Kudo GENKI5, Yoshino RYUNOSUKE6, 7, Hirokawa TAKATSUGU6, 7, Imai KENICHIRO1, 8
1Cellular and Molecular Biotechnology Research Institute, National Institute of Advanced Industrial Science and Technology
2Computational Bio Big-Data Open Innovation Laboratory, National Institute of Advanced Industrial Science and Technology
3Department of Computer Science, School of Computing, Tokyo Institute of Technology
4Middle Molecule IT-based Drug Discovery Laboratory, Tokyo Institute of Technology
5Physics Department, Graduate School of Pure and Applied Sciences, University of Tsukuba
6Division of Biomedical Science, Faculty of Medicine, University of Tsukuba
7Transborder Medical Research Center, University of Tsukuba
8Global Research and Development Center for Business by Quantum-AI Technology, National Institute of Advanced Industrial Science and Technology
( * E-mail: jun.koseki@aist.go.jp )
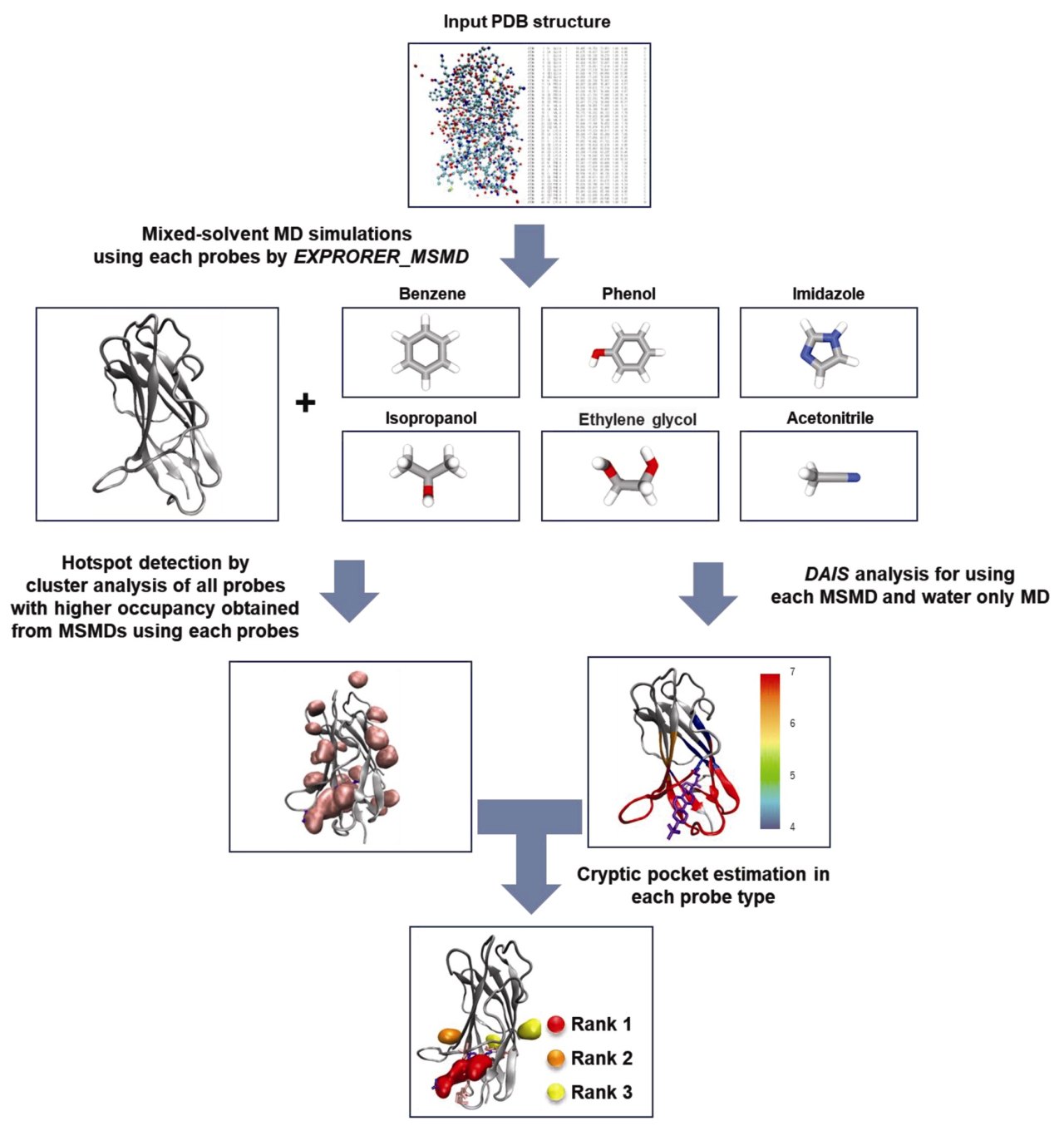
Some functional proteins change their structure to give rise to a binding site only when a binding molecule approaches them. Such binding sites are called cryptic sites and important targets to expand the scope of drug discovery. However, it’s still difficult to predict cryptic sites correctly. Therefore, we propose a method to correctly detect cryptic sites using topological data analysis and mixed-solvent molecular dynamics (MSMD) simulation. To detect hotspots, we employed MSMD simulations using six probes with various chemical properties (Benzene, Isopropanol, Phenol, Imidazole, Acetonitrile, and Ethylene glycol) Then, the possibility of cryptic site was then ranked using our topological data analysis method, the Dynamical Analysis of Interaction and Structural changes (DAIS). For nine target proteins with cryptic sites, the proposed method significantly outperformed the accuracy of the recent machine learning method, Pocketminer. We can detect six of the nine cryptic sites at hotspot Rank 1. In our method, the MSMD simulations with six different probes were employed to search for hotspots showing “ligandability” on protein surfaces, and the DAIS was used for ranking to the possibility of cryptic sites based on estimation of “structural changeability” of protein. The synergistic combination enables to predict cryptic sites with highly accuracy.