P04-02
Dynamical Interaction Energy Analysis of Elastase in Each Reaction State: Insights from Molecular Dynamics and Fragment Molecular Orbital Calculations
Shuhei MIYAKAWA1, Shi Yu TIAN1, Daisuke TAKAYA1, Takayoshi KINOSHITA3, Shigenori TANAKA2, Kaori FUKUZAWA1
1Graduate School of Pharmaceutical Sciences, Osaka University
2Graduate School of System Informatics, Kobe University
3Graduate School of Science, Osaka Metropolitan University
( * E-mail: miyakawa-s@phs.osaka-u.ac.jp )
[Introduction]
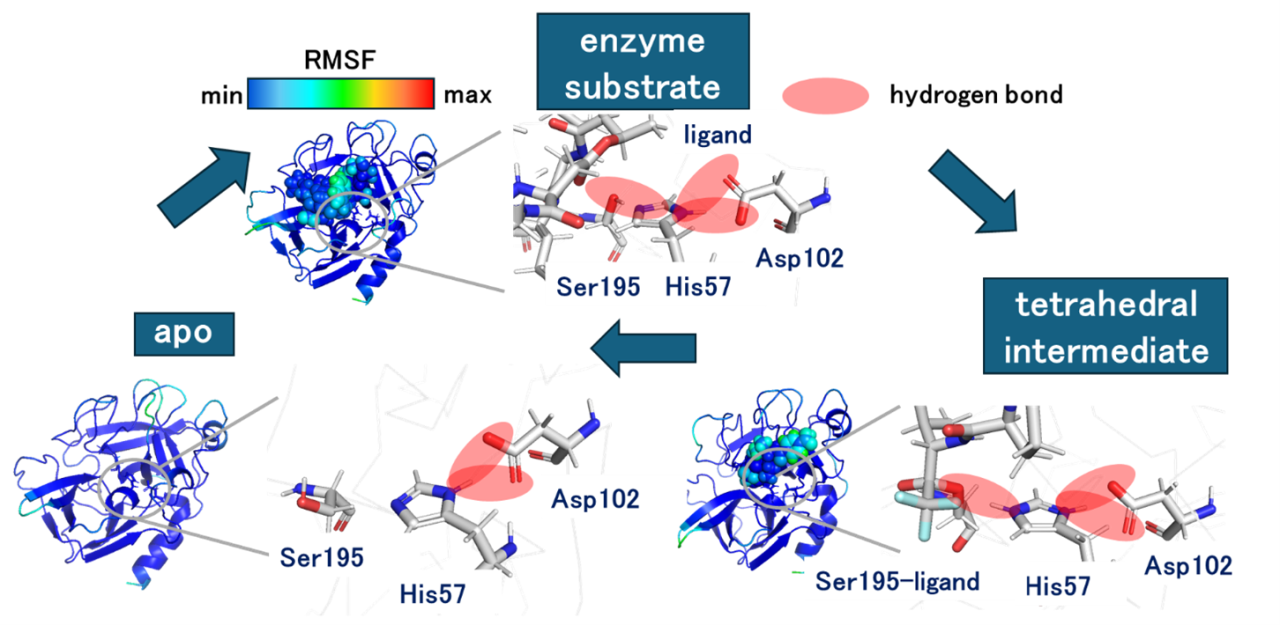
Elastase, a serine protease classified into the chymotrypsin family, has been variously studied in molecular biology and its catalytic triad consists of His57, Asp102, and Ser195 residues. In this study, classical molecular dynamics (MD) and fragment molecular orbital (FMO) calculations were conducted to investigate the dynamical interactions of three states: apo, enzyme-substrate complex and tetrahedral intermediate.
[Methods]
MD calculations by GROMACS were performed for 100 ns × 3 runs on each of the structures: apo and the tetrahedral intermediate structures obtained by neutron crystallography1, and the enzyme-substrate complex structure obtained by X-ray crystallography2, followed by trajectory analysis. FMO calculations were performed on a total of 900 structures extracted from each trajectory at 1 ns interval using the ABINIT-MP program at the MP2/6-31G* level. The inter-fragment interaction energy (IFIE) obtained from the FMO calculation, and pair interaction energy decomposition analysis (PIEDA) were utilized to analyze the hydrogen bond network of the enzyme reaction site. Here, PIEDA component consists of electrostatic energy (ES), exchange repulsion energy (EX), charge transfer energy (CT+mix), and dispersion energy (DI).
[Result & Discussion]
The three reaction states of elastase indicated little structural changes, revealed by the RMSD within 1.8±0.2 Å and the low RMSF values as shown in the figure. However, in the apo state, Ser195, a nucleophilic residue that attacks the substrate, showed in a different conformation from the other states, indicating that the structure of Ser195 did not form a hydrogen bond with His57 in the absence of the substrate. In contrast, PIEDA analysis revealed that the main components of the interaction between Ser195 and His57 were ES and CT+mix, indicating that they formed a stable hydrogen bond in the enzyme-substrate state. The hydrogen bond network at the reaction site was further strengthened in the tetrahedral intermediate state. We will analyze dynamical interactions of the substrates and surrounding residues of elastase for different state of its catalytic cycle.
1. Tamada, T., Kinoshita, T., Kurihara, K., Adachi, M., Ohhara, T., Imai, K., Kuroki, R., & Tada, T. (2009). Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond. Journal of the American Chemical Society, 131(31), 11033–11040. https://doi.org/10.1021/ja9028846
2. Kinoshita, T., Kitatani, T., Warizaya, M., & Tada, T. (2005). Structure of the complex of porcine pancreatic elastase with a trimacrocyclic peptide inhibitor FR901451. Acta Crystallographica Section F, 61(9), 808–811. https://doi.org/10.1107/S1744309105026047