P04-01
Analysis of Kinase Binding Specificity of Staurosporine using the Fragment Molecular Orbital Method
Ruri MIHATA *1, Riko HIGASHINO2, Mayu KITANO3, Shuhei MIYAKAWA1, Shi Yu TIAN1, Daisuke TAKAYA1, Takayoshi KINOSHITA3, Shigenori TANAKA2, Kaori FUKUZAWA1
1Graduate School of Pharmaceutical Sciences, Osaka University
2Graduate School of System Informatics, Kobe University
3Graduate School of Science, Osaka Metropolitan University
( * E-mail: mihata-r@phs.osaka-u.ac.jp )
Introduction
Controlling the selectivity of protein kinases, which are widely distributed in the human body, is a critical issue in drug design. Structural features of kinases, such as the P-loop, essential for ATP binding, and the DFG motif, crucial for the expression of activity, are well known. Staurosporine (STU) exhibits low selectivity and acts as an inhibitor of various protein kinases. This study investigated the detailed interactions between STU and target kinases using computational methods of molecular dynamics (MD) and fragment molecular orbital (FMO), providing insights to understand the binding specificity and selectivity of STU.
Methods
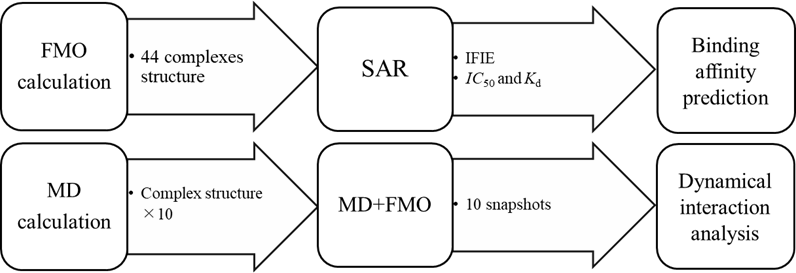
Employing complex structures of STU and 44 kinds of kinases from Protein Data Bank (PDB) based on X-ray crystallography, FMO calculations were conducted to analyze Inter-Fragment Interaction Energy (IFIE). Structure-activity relationships (SAR) between IFIE and kinase inhibitory activity IC50, as well as kinase binding activity Kd, were investigated. In addition, ten MD runs of 100 ns between ALK kinase and STU were performed. For ten snapshots from each MD trajectory, FMO calculations were performed (MD+FMO). MD calculations were performed using GROMACS under ff14SB for protein force field, TIP3P for water force field, GAFF2 for ligand force field and FMO calculations were conducted using ABNIT-MP at a calculation level of MP2/6-31G*. The calculation flow is shown in fig.
Results and Discussion
The SAR analysis was performed on 44 different kinases with low correlation coefficient (R² = 0.17), and classification of kinases based on P-loop and DFG-motif did not improve the correlation. Therefore, to remove the influence of crystal packing, MD calculations were performed. The MD results showed that while the RMSD of the overall structure was suppressed to 2.05±0.22 Å, the RMSD of the P-loop was large at 2.95±0.77 Å, suggesting that the structure was relaxed in an aqueous environment. Additionally, a comparison was made between the IFIE from the crystal structure and the dynamically averaged IFIE from MD+FMO. A significant difference was observed in the interaction between STU and Asp1203, where the IFIE from the crystal structure was -106.1 kcal/mol, while the averaged IFIE from MD+FMO was -43.5 kcal/mol. These findings suggest that crystal packing affects the interaction energy of STU and kinase, and comprehensive MD+FMO study is a promising approach to understanding kinase binding specificity.