P03-30
Directional Graph Modelling for Solution Design and Experiment Automation
Yusuke SAKAI *
Center for Biosystems Dynamics Research, RIKEN
( * E-mail: yusuke.sakai@riken.jp )
The popularisation of smart analytical equipment and liquid handling devices has made laboratory automation crucial for enhancing research efficiency. Automation advances science by improving reproducibility, enabling high-throughput investigations, and enhancing work-life balance. Research automation now spans experimental procedures, experiment design, record-tracking, data analysis, and interpretation.
This presentation focuses on automating experimental stages in DNA nanotechnology and synthetic biology, showcasing an in-house relational database (RDB) solution and automated liquid handling devices. DNA origami, which involves hundreds of components, demands laborious mixing and careful inventory management of numerous oligo DNAs. As research advances, the number of DNAs can tens of thousands, with countless combinations. To address these challenges, we developed a tool called ‘SolutionDesigneRDB (SDRDB)’ to automate these processes, reducing reliance on researchers’ diligence.
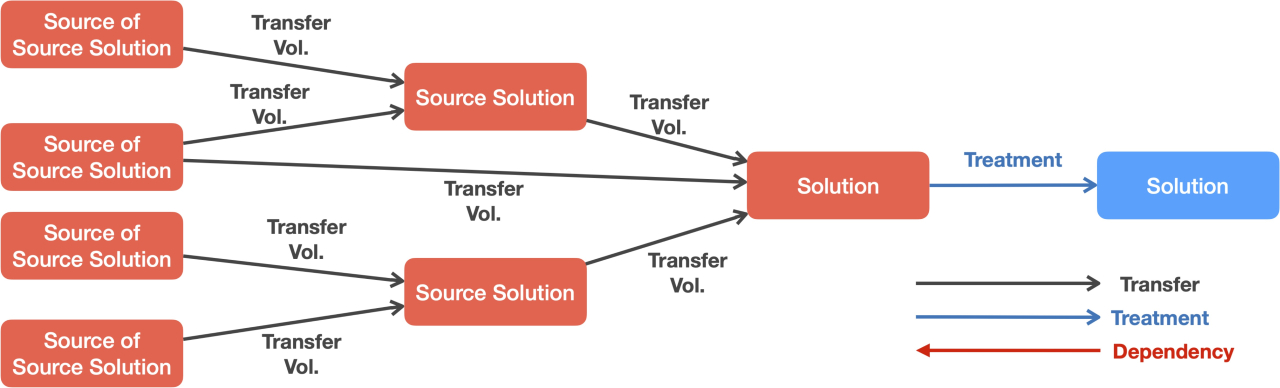
SDRDB, build using Claris FileMaker, is a low-code, GUI-rich relational database application. The solution design within the app is modelled as a directed graph, where each node represents an individual solution, and the directional edges represent the volume of liquid transferred. The sample preparation processes, such as liquid handling, heating, and separating, are similarly modelled, with each node representing a specific state of an individual solution, and the edges showing their dependency (process order) and procedural details. The main features of the app include: (1) an RDB that supports multi-stage mixing and essential experimental procedures, allowing systematic and dynamic configuration of sample preparation, (2) a user-friendly GUI for designing solution compositions within the database, either in batches or individually, (3) scripts to generate protocol files for specific liquid handling devices, and (4) scripts for importing oligo DNA lists generated by cadnano into the database for each DNA origami structure (the primary goal of the system). Due to its low-code design, users can easily adapt scripts and the GUI to fit their specific research needs with minimal effort. SDRDB’s solution design and integration with liquid handling systems help overcome bottlenecks in research involving complex mixture designs, not just in DNA nanotechnology.
We are seeking funding and collaborators to migrate the system to a free web application using PostgreSQL or equivalent and JavaScript for broader accessibility. We also aim to re-implement the system in a native Graph Database model, renaming it SolutionDesiGnerDB (SDGDB) for enhanced data analysis, or NoSQL (SolutionDesigNerDB, SDNDB) for multimodal extension. Additionally, we intend to integrate Bayesian inference functions for automatic sample series formulation, upgrading the app to ExperimentDesigneRDB (EDRDB).