P03-12
Deep learning-based enzyme screening to identify orphan enzyme genes
Keisuke HIROTA *, Takuji YAMADA
School of Life Science and Technology, Institute of Science Tokyo
( * E-mail: hirota.k.221@gmail.com )
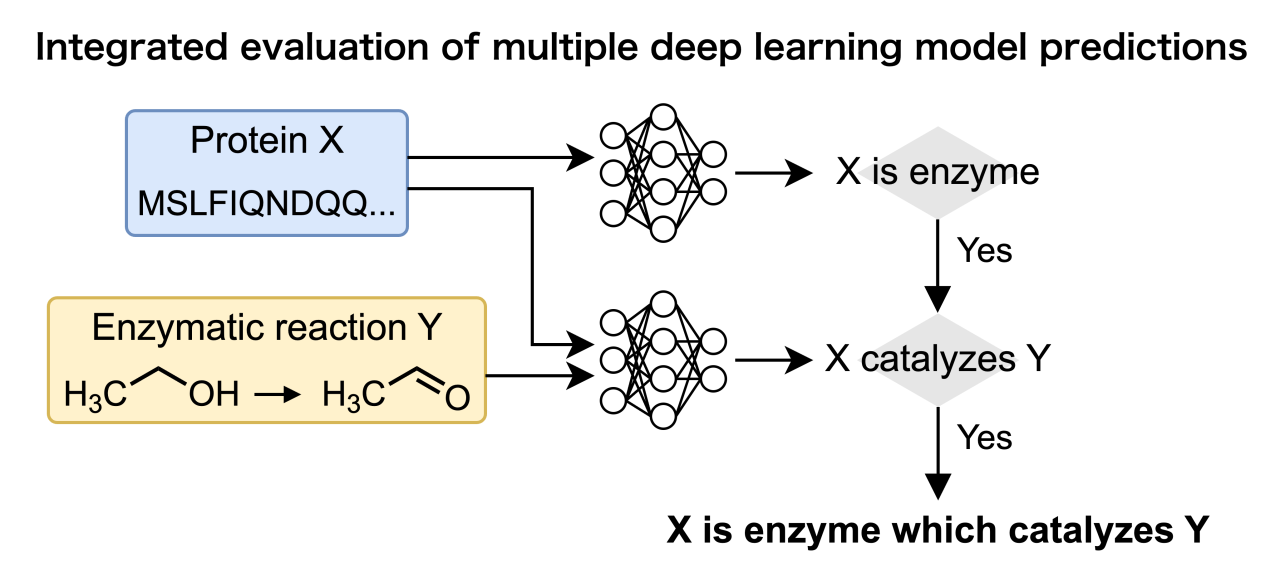
Progress in sequencing technology has yielded large numbers of protein sequences, including enzymes which are important proteins that catalyze specific chemical reactions. However, many proteins of unknown function are encoded in genomes from bacteria to humans, and many functions of living organisms remain unsolved. As for enzymes which are important proteins that catalyze specific chemical reactions, many enzymes are without sequence information in databases. Those enzymes whose activities are known but whose sequences are unknown are called orphan enzymes. This gap between known proteins and enzyme reactions suggests that some proteins of unknown function might be orphan enzymes. However, most existing tools to predict enzymatic function rely on EC numbers and have limited applicability to orphan enzymes, which are often not sufficiently annotated with EC numbers. Moreover, previous studies that assign protein sequences to enzymatic reactions based on reaction similarity cannot handle proteins of unknown function. Therefore, a computational tool is needed to evaluate the correspondence between any enzyme reaction and protein sequence before costly experimental validation.
In this study, we propose a deep learning-based framework for enzyme screening that takes protein sequences and enzyme reactions as inputs and evaluates their correspondence. The proposed method differs from previous studies in two aspects. First, our approach performs a two-step evaluation: classification of enzymes and non-enzymes to handle any given protein including non-enzyme proteins, and prediction of their correspondence with enzymatic reactions of interest. Second, the proposed method embeds enzymatic reactions into vector representations, enabling it to handle reactions that cannot be annotated with enzymatic reaction labels, such as EC numbers. This study then aims to establish a computational tool to evaluate the correspondence between any given enzymatic reaction and protein sequence and to enable a fast and large-scale search for orphan enzyme candidates.