P03-05
Computational determination of SMARTS molecular query containment relationships
Seiji MATSUOKA *1, Minoru YOSHIDA1, 2, 3
1Center for Sustainable Resource Science, RIKEN
2Office of University Professors, The University of Tokyo
3Collaborative Research Institute for Innovative Microbiology, The University of Tokyo
( * E-mail: smatsuoka@riken.jp )
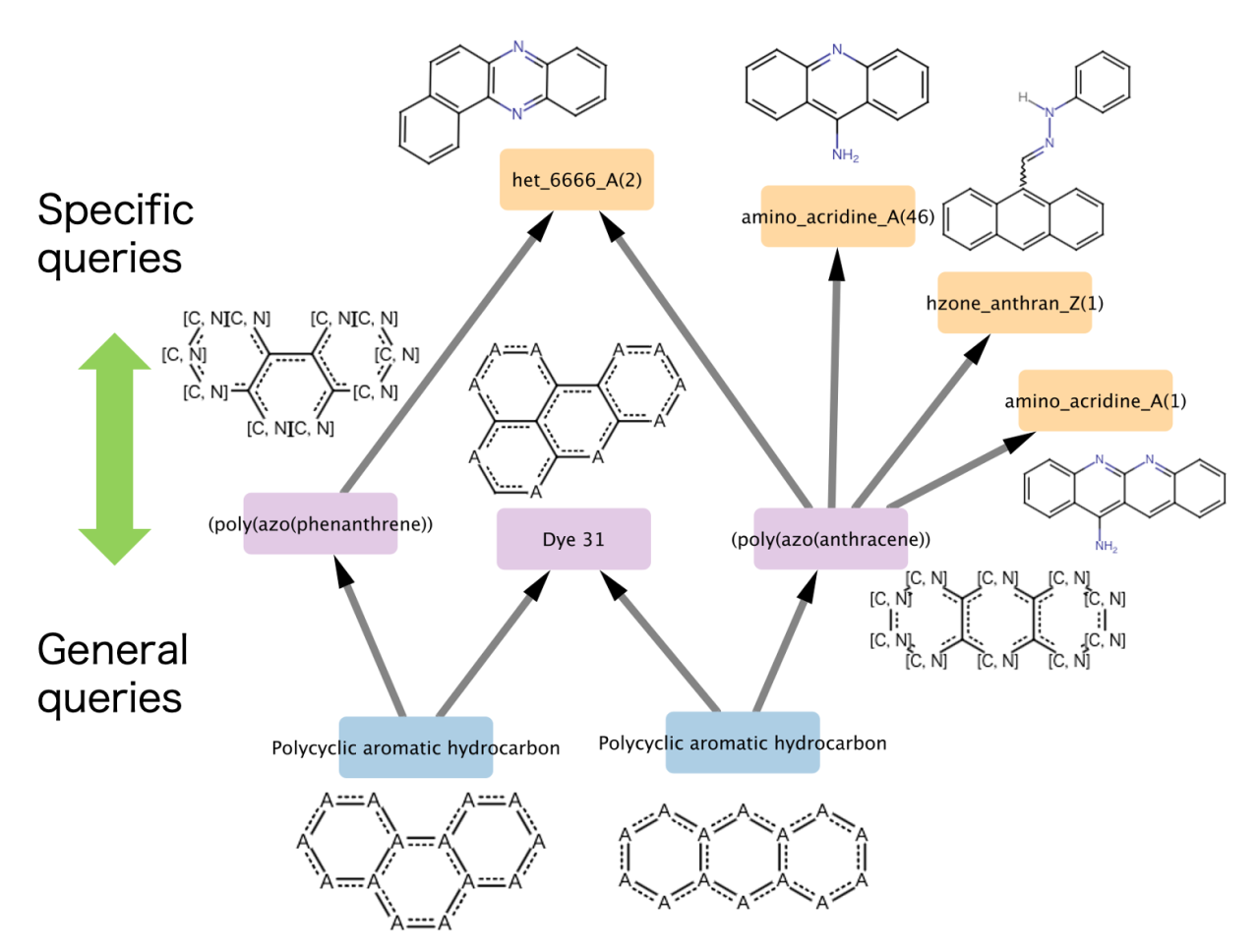
Describing molecules as substructure patterns is convenient for focusing on particular substructures and functional groups that are responsible for a distinctive function of the molecules. For example, patent claims often cover a group of compounds that meet certain criteria, expressed as substructure patterns called Markush structures. A database of substructure patterns in a machine-readable format is promising for chemical information systems, such as those used for patent or regulatory chemical searches. However, this involves significant challenges. To search substructure patterns using a substructure pattern query, it is necessary not only to match substructures but also to determine the containment relationships of each atom/bond attributes, including logical expressions.
We have developed an algorithm for determining containment relationships of SMARTS, a commonly used molecular query language. This algorithm consists of a substructure matching based on the VF2 algorithm and containment relationship determination of atom/bond attributes by using truth tables that can handle logical operators and recursive expressions in SMARTS. This is implemented as part of MolecularGraph.jl, a molecular graph modeling toolkit, and is available as open-source software.
This method also allows a substructure pattern dataset to be represented as a directed acyclic graph (DAG) by obtaining containment relationships for all combinations of substructure patterns. This capability was demonstrated through the systematic visualization of relationships in ChEMBL structural alerts dataset used in medicinal chemistry. Additionally, a dataset of frequently occurring functional groups and substructures was constructed to generate containment relationships in a DAG. This enables the functional annotation of molecules as a subgraph of that DAG. Such an approach can potentially be applied to the development of molecular descriptors that capture features of molecular functions.