P03-04
Reaction-Aware Molecular Optimization Using Conditional Transformer and Reinforcement Learning
Shogo NAKAMURA *1, Nobuaki YASUO2, Masakazu SEKIJIMA3
1Department of Life Science and Technology, Tokyo Institute of Technology
2Academy for Convergence of Materials and Informatics (TAC-MI), Tokyo Institute of Technology
3Department of Computer Science, Tokyo Institute of Technology
( * E-mail: nakamura.s.bu@m.titech.ac.jp )
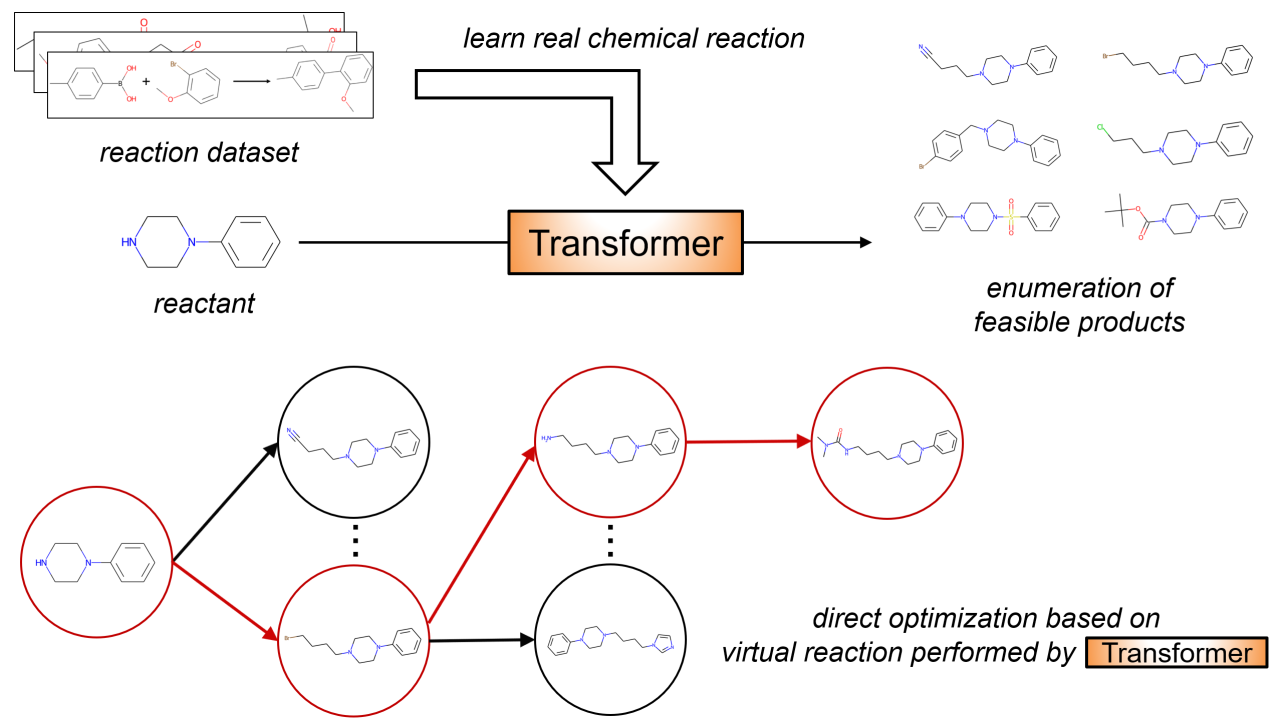
Designing molecules with desirable properties is an important research issue in drug discovery. Recent advances in deep learning have led to the development of molecular generation models to obtain compounds with desired properties. However, existing compound discovery models often ignore the key issue of ensuring the feasibility of organic synthesis. To address this issue, we propose TRACER (molecular optimization using a conditional Transformer for reaction-aware compound exploration with reinforcement learning). The core of TRACER is a Conditional Transformer model trained on a dataset of chemical reactions. By explicitly training on chemical reactions, the model can predict realistic products from a given reactant under the constraints of the reaction type specified by the graph convolutional network. Molecular optimization results in the activity prediction model targeting the dopamine receptor D2 showed that TRACER effectively generated compounds that scored highly. The structure-wide Transformer model captures the complexity of organic synthesis and allows exploration within the vast chemical space while accounting for real-world reactivity constraints The source code, activity prediction models, and curated datasets are available in our public repository (available at https://github.com/sekijima-lab/TRACER).