P02-06
SpatialPPI 2.0: Enhancing Protein-Protein Interaction Prediction Through Distance Matrix Analysis Using Link Regression in Graph Attention Networks
WENXING HU *, Masahito OHUE
Tokyo Institute of Technology
( * E-mail: perseids2032@gmail.com )
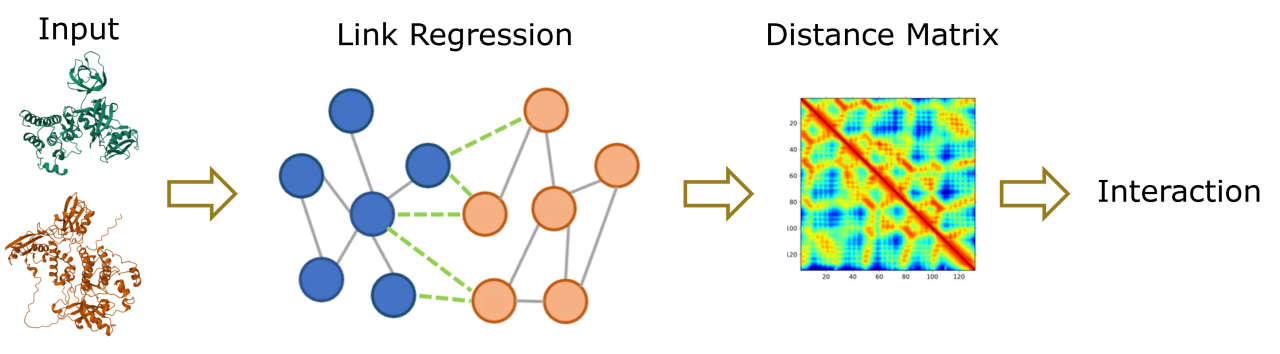
Protein-protein interactions (PPIs) play a pivotal role in a wide array of biological processes, and accurately predicting these interactions is essential for advancing our understanding of cellular functions. In this study, we introduce a novel computational approach that leverages link regression within Graph Attention Networks (GATs) to predict spatial distances between two independent protein structures. Our method is designed to enhance the prediction of PPIs by focusing on the distance between residues rather than the conventional approach of contact map prediction.
We trained our model on the Protein-Protein Interfaces Prediction dataset from the ATOM3D project and validated its effectiveness on AlphaFold-predicted datasets. The uniqueness of our approach lies in its utilization of link regression to estimate inter-residue distances, which can be further processed to refine the prediction of protein-protein interfaces. Unlike existing methods that employ link classification, our model directly predicts the distance between residues, providing a more nuanced understanding of potential interaction sites.
Traditional GNN-based models typically use a Siamese network to independently extract features from each protein structure. In contrast, our method innovatively combines two protein structures with variable links between them, allowing the model to dynamically update the distances and capture the influence of residues across the interacting proteins. This approach offers a more holistic view of protein interactions, as it considers the complex interdependencies between residues in both proteins.
Comparative analysis with existing methods such as D-SCRIPT, which uses sequence-based approaches and contact maps as intermediate predictions, demonstrates the potential of our model to improve the accuracy and reliability of PPI predictions. Our method also shows promise when compared to other GNN-based models like PIPR and GNNGL-PPI, highlighting the advantages of integrating spatial distance prediction with graph-based learning.
The potential applications of our findings are far-reaching, extending to any domain where accurate PPI prediction is critical. This includes drug discovery, where understanding PPIs can inform the design of inhibitors or enhancers, and in systems biology, where mapping interaction networks can elucidate the pathways underlying various diseases. This study underscores the potential of GAT-based models with link regression in advancing the field of structural bioinformatics, offering a new direction for PPI prediction methodologies.