P02-03
Fragment Molecular Orbital Calculations for Zinc-Containing smHDAC8
Siyun WANG *, Sota TANAKA, Shuhei MIYAKAWA, Yu-Shi TIAN, Daisuke TAKAYA, Kaori FUKUZAWA
Graduate School of Pharmaceutical Sciences, Osaka university
( * E-mail: wangsiyun001121@outlook.com )
Background
Fragment Molecular Orbital (FMO) calculations have been proven useful in drug design and protein-ligand binding analysis. However, metal ions contained in biomolecule systems limit the application of this method due to the difficulty in both fragmentation and complex electronic structure of transition metal ions. This study used a zinc (Zn²⁺)-containing protein Schistosoma mansoni Histone Deacetylase 8 (smHDAC8) to investigate the methodology and attempted to provide a protocol for FMO calculation for metalloprotein.
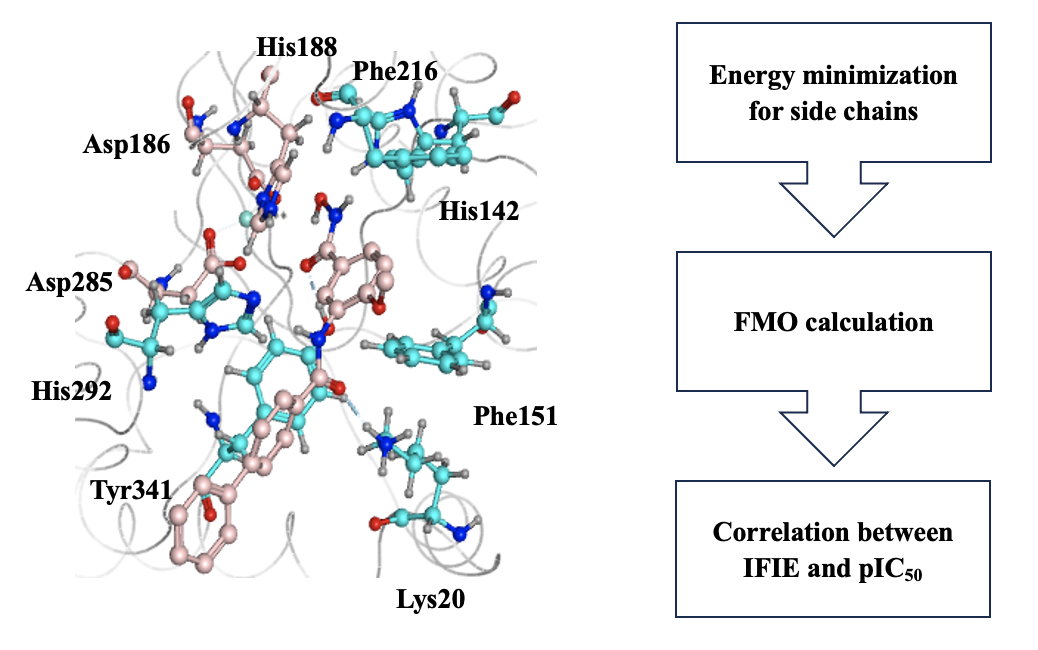
Methods
smHDAC8 contains a catalytic Zn²⁺ active site for ligand binding. 14 active ligands of smHDAC8 have been reported by Martin Marek et al. [1]. Initially, we selected one complex as a template structure (PDB ID: 6hsh). Other complexes were constructed by replacing the coordinates of ligands and performing structural optimization using Amber10:EHT force field. All the structures were prepared by MOE. After structure preparations, energy minimizations for side chains were conducted using a constraint of tether 1.0. Subsequently, Zn²⁺ and surrounding residues were constructed as one large fragment (i.e., merging code). FMO calculations were performed using the 6-31G* basis set . The energy analysis was conducted in a “super-molecule” manner:
Ebind=Eprotein+Eligand-Ecomplex.
Finally, the binding energies of ligands were compared with their biological activities measured as IC50 values.
Results and Discussion
FMO calculations for all these complex models succeeded, indicating an executable approach for metalloprotein. The interactions between the key ligand and the protein observed in the experiment were validated using IFIE analysis performed with FMO, and the results were consistent. Additionally, it was found that the residues His142, Phe151, and Phe216 form π-π interactions with the ligand, which are also crucial for the ligand function.
Reference
[1] Marek M, Shaik TB, Heimburg T, et al. Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants. J Med Chem. 2018;61(22):10000-10016. doi:10.1021/acs.jmedchem.8b01087