P02-02
PairMap: An Intermediate Insertion Approach to Improve Accuracy in Relative Free Energy Perturbation Calculations of Distant Compound Transformations
Kairi FURUI *1, Takafumi SHIMIZU2, Yutaka AKIYAMA1, Roy S. KIMURA2, Yoh TERADA2, Masahito OHUE1
1School of Computing, Institute of Science Tokyo
2Alivexis, Inc.
( * E-mail: furui@li.c.titech.ac.jp )
Accurate prediction of the difference in binding free energy between compounds is crucial for reducing the high costs associated with drug design and lead optimization.
Relative binding free energy perturbation (RBFEP) calculations are effective for small structural changes; however, large topological changes pose significant challenges for calculations, leading to high errors and difficulties in convergence.
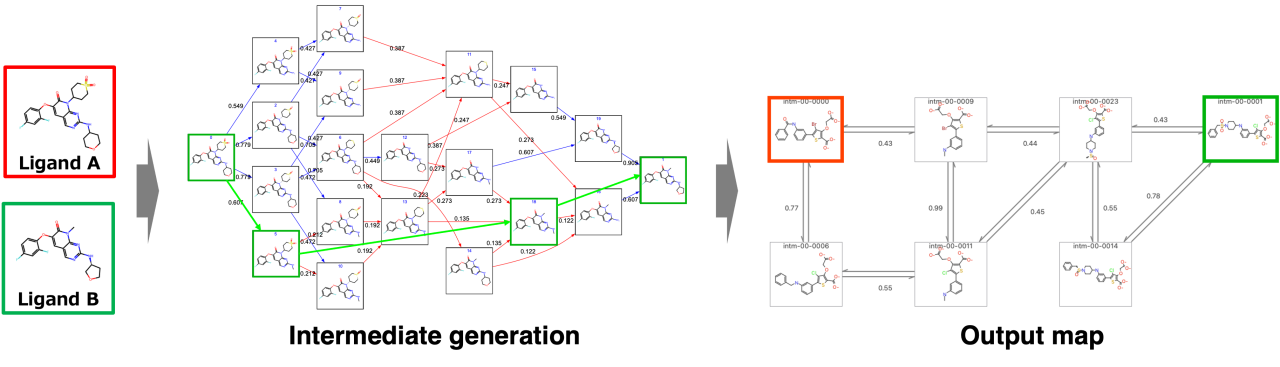
To address such issues, we propose a new approach---PairMap---that focuses on introducing appropriate intermediates for complex transformations between 2 compounds. PairMap generated intermediates exhaustively, determined the optimal conversion paths, and introduced thermodynamic cycles into the perturbation map to improve accuracy and reduce computational cost.
PairMap succeeded in introducing appropriate intermediates that could not be discovered by existing simple approaches by comprehensively considering intermediates.
Furthermore, we evaluated the accuracy of the prediction of binding free energy using 9 compounds selected from Wang et. al. 's benchmark set, which included particularly complex transformations. The perturbation map generated by PairMap achieved excellent accuracy with a mean absolute error of 0.93 kcal/mol compared to 1.70 kcal/mol when using the perturbation map generated by the conventional Flare FEP intermediate introduction method.
Moreover, in a scaffold hopping experiment conducted with the PDE5a target involving complex transformations, PairMap provided more accurate free energy predictions than ABFEP calculations, yielding more reliable results compared to experimental data.
Additionally, PairMap can be utilized to introduce intermediates into congeneric series, demonstrating that complex links on the perturbation map can be resolved with minimal addition of intermediates and links.
In conclusion, PairMap overcomes the limitations of existing methods by enabling RBFEP calculations for more complex transformations, further streamlining lead optimization in drug discovery.