P01-19
Molecular simulation analysis for nucleic acids
Kenji YAMAGISHI *1, Hiroyuki TSUKADAH2, Haruto NARITA1, Shota MYOCHIN1, Hisae YOSHIDA1, Seiichiro ISHII1, Masahiro SEKIGUCHI1, Takeshi ISHIKAWA3, Taiichi SAKAMOTO4
1Nihon University
2Nissan Chemical CORPORATION
3Kagoshima University
4Chiba Institute of Technology
( * E-mail: yamagishi.kenji@nihon-u.ac.jp )
A nucleic acid is a macromolecule composed of nucleotide chains. The most common nucleic acids, deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), are biopolymers essential to life. We studied the structure and function of the nucleic acids using molecular simulations. In this presentation, we provide an overview of our research project.
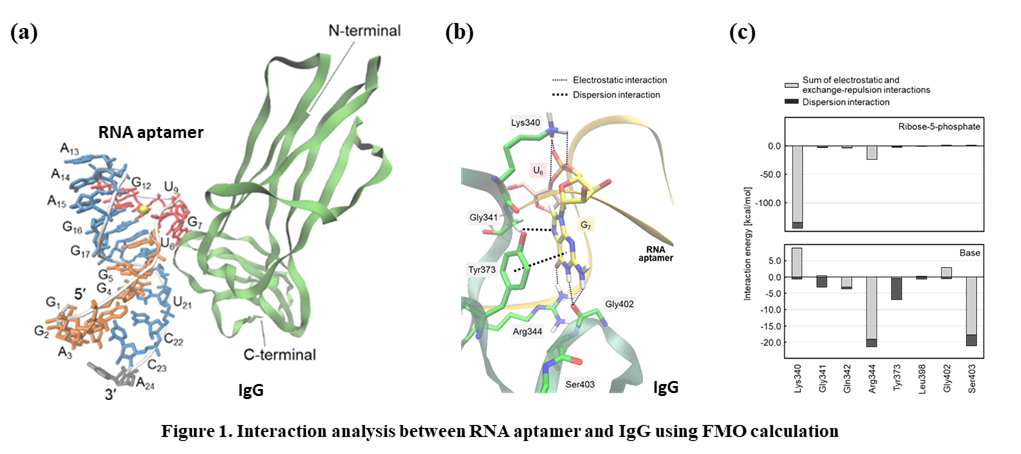
1. Aptamer Aptamers are short, single-stranded oligonucleotides that bind to specific target molecules such as proteins, nucleic acids, and small molecules. An aptamer that binds to the Fc portion of human Immunoglobulin G (IgG) was identified using the SELEX technique. We have been conducting research aimed at clarifying the mechanism of the high affinity and specificity of the aptamer against IgG using fragment molecular orbital calculations and molecular dynamics simulations.
Ref [1] H. Yoshida, T. Ishikawa, T. Sakamoto, K. Yamagishi, et al., Chem. Phys. Lett., 738, 136854 (2020).
2. Antisense Oligonucleotides Antisense oligonucleotides (ASOs), composed of single-stranded DNA-like oligonucleotides, represent a key category of oligonucleotide therapeutics, and constitute the majority of oligonucleotide therapeutics approved by 2024. We have been conducting molecular dynamics simulations of the RNase H complex with ASO/RNA duplex structure to elucidate the structural dynamics and interactions.
3. Modified Nucleoside Various chemically modified nucleoside analogs have been developed to improve the properties and performance of natural nucleic acids. We performed molecular dynamics calculations of the novel thymidine analogs to investigate the changes in nucleoside sugar puckering and molecular fluctuations caused by chemical modifications.
Ref [2] Y. Zhou, S. Ishii, K. Yamagishi, Y. Ueno, et al., RSC Adv., 10, 41901 (2020).