P01-11
The Computational Study on the Secondary Structure Formation of Nascent Peptides Inside the Ribosome Tunnel with Biomolecular Environments Mimicking Model
Takunori YASUDA *1, Rikuri MORITA2, Yasuteru SHIGETA2, Ryuhei HARADA2
1Doctral Program in Biology, Institute of Life and Environmental Sciences, University of Tsukuba
2Center for Computational Sciences , University of Tsukuba
( * E-mail: takunoriyasuda@gmail.com )
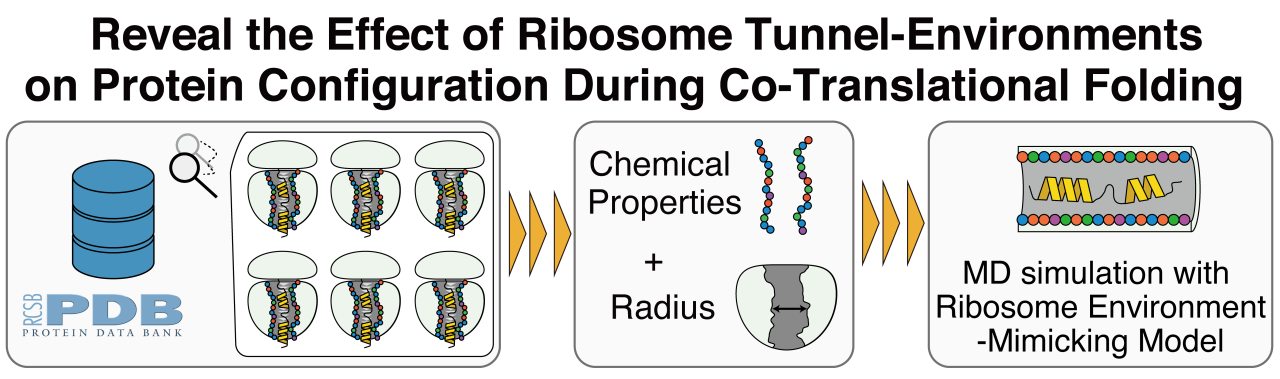
The conformation of proteins is determined not only by their sequence but also by surrounding environment. Therefore, investigating the conformations in the cellular environment is crucial for understanding protein function in cells. To investigate protein configurations within diverse biomolecular environments, several approaches that combine molecular dynamics (MD) simulations with simplified models have been proposed. Specifically, carbon nanotubes and hydrophobic cages have been well established as models of ribosome-tunnels or protein chaperons, respectively. However, recent findings suggest that these uniform hydrophobic models may not adequately capture the effects within each biomolecular environment. Based on these facts, it is necessary to generate spherical and cylindrical models based on a variety of chemical properties corresponding to the components within target biomolecules. Therefore, we developed a new open-source tool called Biomolecular Environment-Mimicking Model Generator (BEMM-GEN).
Furthermore, as an application of BEMM-GEN, we focused on the ribosomal tunnel. Inside the ribosome tunnel, a nascent peptide forms its specific α-helix. Despite the deeply relationship between such temporally α-helix formation and biological functions, a comprehensive analysis of the ribosome tunnel environment’s impact on the α-helix formation has not been conducted yet. Therefore, we generated a computational model called Ribosome Environment-Mimicking Model (REMM) by considering the radius and components of experimentally determined ribosome structures. Using an enhanced all-atom molecular dynamics simulation, we investigated the properties of the nascent peptides in the experimental structures inside the Carbon nanotube (CNT), in addition to the REMM. Herein, we adopt the CNT as a reference model and compared the ability of both models to replicate the α-helix of the nascent peptides. Finally, we elucidated the mechanism of ribosome tunnel-specific α-helix formation by analyzing the nascent peptides inside each model.