P01-08
Comprehensive docking simulations using AlphaFold2-based human olfactory receptors for odor prediction
Hirotada KANESHIRO *1, Masakazu SATO1, Airi TANAKA1, Shuya NAKATA1, Yoshiko AIHARA2, Hirotaka Nishioka KITOH3, Yoshiharu MORI4, Shigenori TANAKA1
1Laboratory of Computational Science, Graduate School of System Informatics, Kobe Univ., Kobe, Japan
2Grad. Sch. of Agri. Sci., Kobe Univ., Kobe, Japan
3Fac. of Sci. and Eng., Kindai Univ., Osaka, Japan
4KQCC, Keio. Univ., Yokohama, Japan
( * E-mail: 238x017x@stu.kobe-u.ac.jp )
When we smell, we can distinguish many odors, but it is difficult to predict an odor from its molecular structure information.
The fact that there is no clear classification of smells, such as the five tastes in the sense of taste, also makes it difficult to predict smells.
Therefore, the purpose of this study is not to make absolute odor prediction, but to predict which odor molecules outside the database are closest to which molecules in the database by using odor molecules and odor descriptors registered in the database ATLAS.
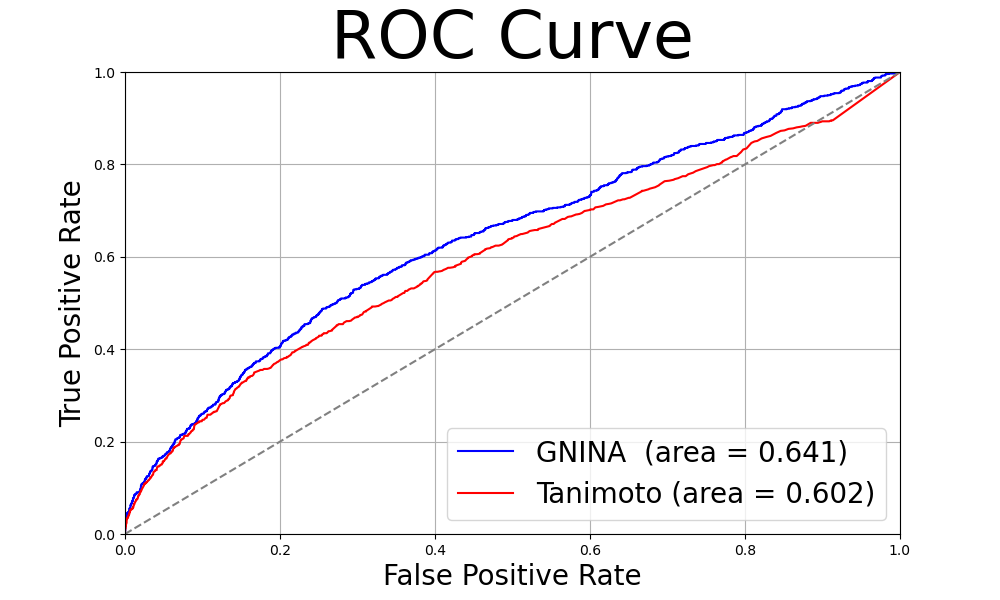
One simple approach to this comparison of odorant molecules is to compare the structural similarity of odorant molecules, but in this study, odor similarity is examined based on docking scores with human olfactory receptors.
Specifically, using OR52c (PDBID:8HTI), which has recently been structurally determined by cryo-EM, as a template, we generated approximately 400 human olfactory receptor structures using AlphaFold2 and performed comprehensive docking using GNINA with approximately 130 odor molecules from ATLAS.
The scores obtained at this time were compared with a score (Tanimoto coefficient) that expresses the structural similarity of the molecules.
the results were found to be superior to those obtained using an index that evaluates the similarity of molecular structure.
The results are shown in the following figure.