P01-06
Enhanced Prediction of Antigen-Antibody Complex Structures through Aggressive Structural Refinement by AlphaFold2
Seiya TANAKA *1, Masaki KOYAMA1, Hiroki ONODA2, Leonard CHAVAS1, 2, George CHIKENJI1
1Department of Applied Physics, Graduate School of Engineering, Nagoya University
2Synchrotron Radiation Research Center, Nagoya University
( * E-mail: tanaka.seiya.a1@s.mail.nagoya-u.ac.jp )
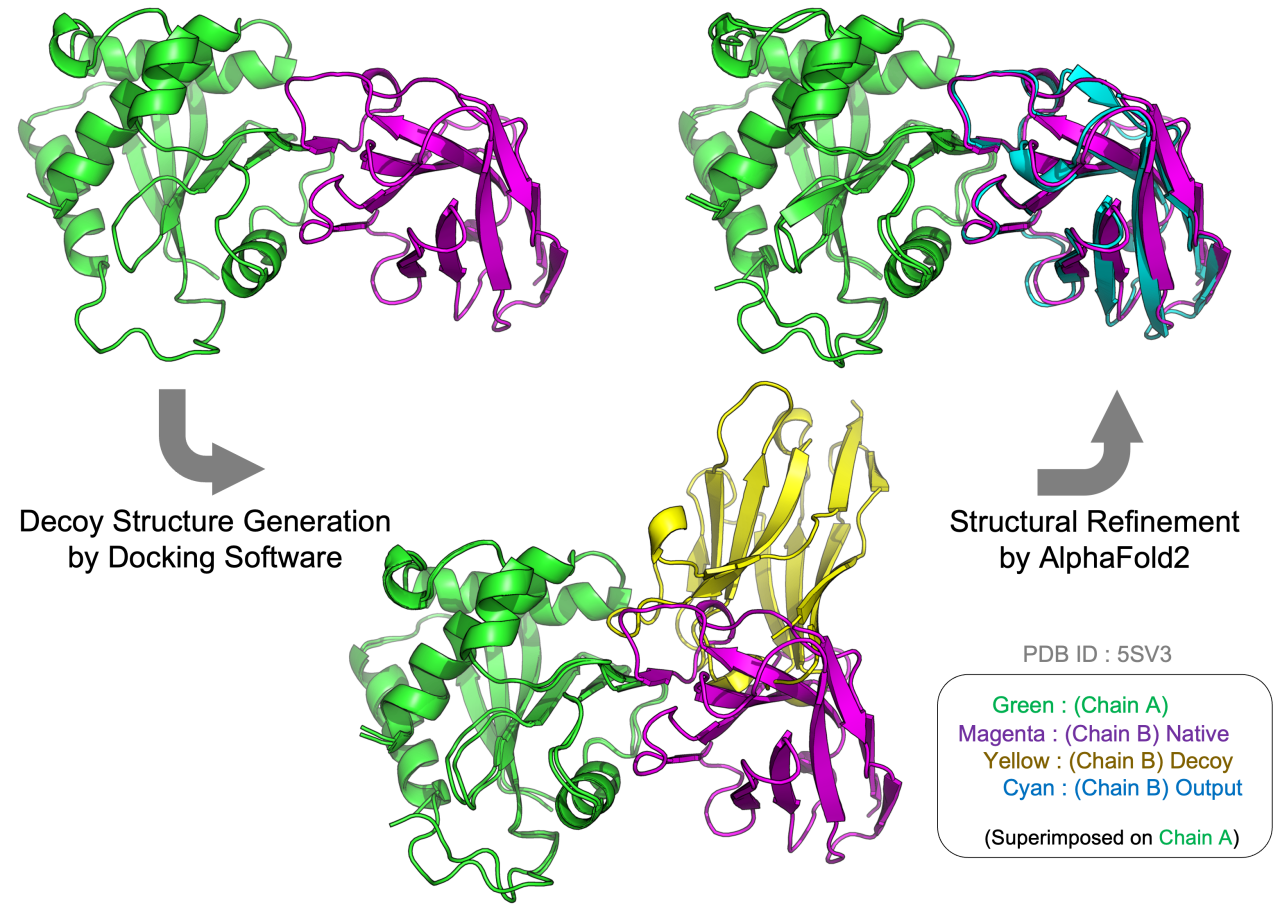
Accurate prediction of protein structures is crucial for elucidating their biological functions. AlphaFold2, a cutting-edge machine learning neural network algorithm, has significantly enhanced the accuracy of monomer structure predictions by leveraging co-evolutionary information. However, predicting protein complex structures, particularly Antigen-Antibody complexes, remains a formidable challenge largely due to the absence of co-evolutionary signals at the interaction interfaces. Recent progress has integrated AlphaFold2 with physics-based docking programs to address this gap, leading to success rates that surpass those observed with AlphaFold2 alone in complex structure prediction. Despite these advancements, success rates have considerable room for improvement. The current study introduces a methodology enhancing the precision of complex structure predictions. Our approach employs a physics-based protein docking program to generate multiple decoys for input into AlphaFold2 for quality refinement and confidence evaluation. A novel aspect of our method is the modification of AlphaFold2 to allow aggressive structural refinement. We demonstrate that our approach enables significant conformational adjustments in certain complexes, achieving closer proximity to native structures. This structural refinement performed by AlphaFold2 improves the accuracy of predicting complex structures, particularly for Antigen-Antibody complexes, making a significant methodology advancement.