P01-01
Hepatitis C Virus Drug Resistance Mechanism: Docking and Molecular Dynamics Study of NS5A-Drug Complex
Yaxuan WANG*1, Ai TOYODOME 2, Seiichi MAWATARI 2, Midori TAKEDA 3, Masanori IKEDA 3, Takeshi ISHIKAWA 1
1 Graduate School of Science and Engineering, Kagoshima University
2 Graduate School of Medical and Dental Sciences, Kagoshima University
3 Joint Research Center for Human Retrovirus Infection, Kagoshima University
( * E-mail: k4340100@kadai.jp )
Chronic hepatitis C virus (HCV) infections can lead to hepatocellular carcinoma, and the virus's high genotypic variability hinders effective vaccine development. Inhibitors against the NS5A protein, such as Daclatasvir, Ledipasvir, Velpatasvir, Elbasvir, and Pibrentasvir, are used as antiviral agents. However, they are highly resistant to Q24K/L28M/R30Q/A92K mutations, with Pibrentasvir showing resistance to R30E but not other mutations[1,2]. In this study, we modeled the HCV NS5A protein structure to understand the resistance mechanism of these antiviral agents.
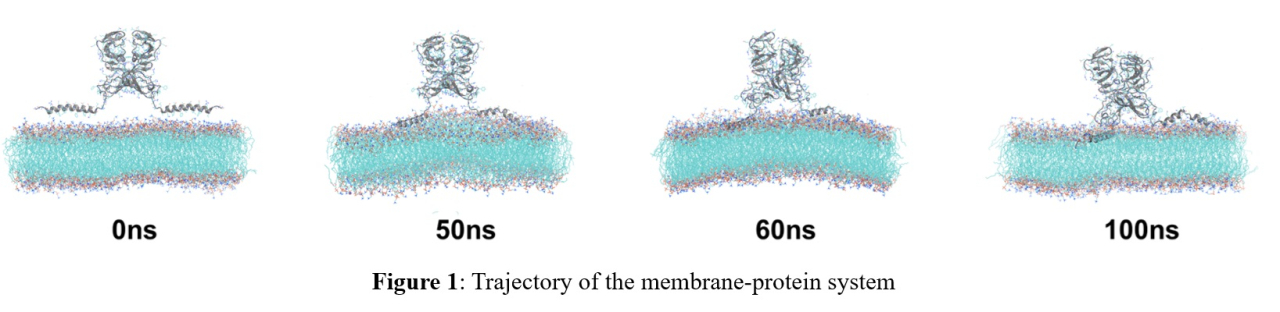
Our study focuses on the N-terminal drug resistance mutation site of the NS5A protein. Since no crystal structure, including this mutation site, has been reported, homology modeling was performed to create a three-dimensional structure model, and then docking simulations were performed using AutoDock Vina[3] to create complex models with the inhibitors. However, our models failed to adequately explain the drug resistance mechanism. Next, to mimic the human body's environment more closely, we attempted molecular dynamics (MD) simulations for NS5A protein with biological membranes. The genmixmem program[4] was used for modeling, and GROMACS[5] was used for MD simulations. A 100 ns trajectory of the membrane-protein system was obtained (Figure 1) and the inhibitors attached to the 60 ns structure, by which membrane-protein-inhibitor systems were created. Now, we are proceeding with the MD simulations for these systems.
[1] R. Bartenschlager, et al., The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection, Nature Reviews Microbiology, 11 (2013) 482.
[2] A. Toyodome, et al., Analysis of the susceptibility of refractory hepatitis C virus resistant to nonstructural 5A inhibitors, Sci. Rep., 14 (2024) 16363.
[3] O. Trott and A. J. Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem., 31 (2010) 455.
[4] T. Lu, J. Wang, and L. Xu, genmixmem program (http://sobereva.com/245).
[5] M. J. Abraham, et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software X, 1-2 (2015) 19.