O07_06
Deep Learning-Based Protein-Protein Interaction Prediction Considering Angle Information
Yukinobu MATSUNO *, Masakazu SEKIJIMA
Department of Computer Science, Tokyo Institute of Technology
( * E-mail: matsuno.y.af@m.titech.ac.jp )
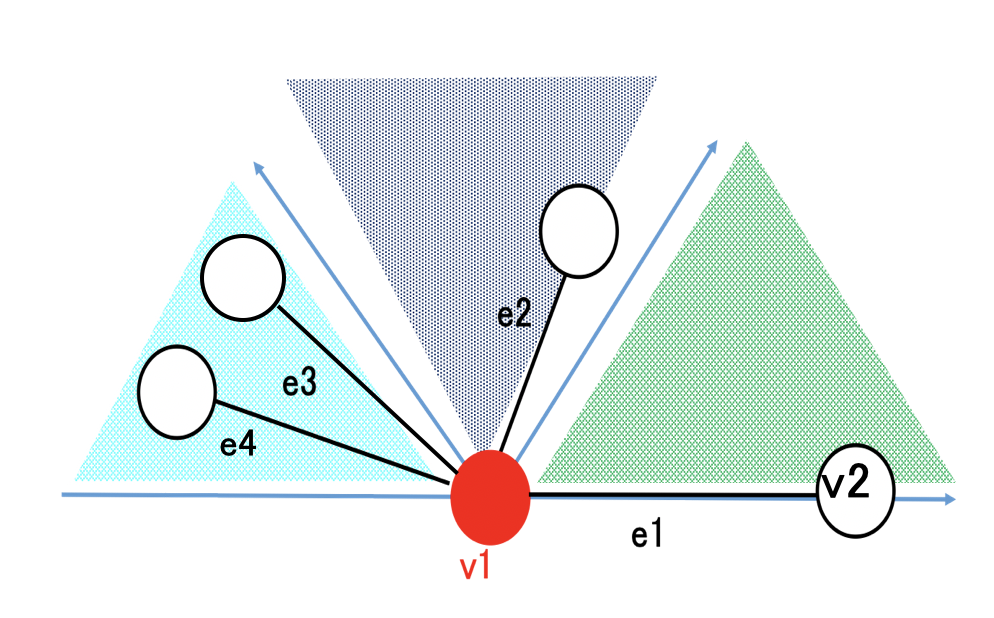
Many proteins are known to control cellular functions through protein-protein interactions (PPIs). Therefore, understanding the structure of protein complexes is valuable information for structure-based drug discovery. Although experimental methods such as X-ray crystallography exist, determining the structure of protein complexes requires enormous costs and expenses. As a result, computational methods called docking have been developed to predict the structure of protein complexes. Evaluating the structures obtained by docking using only the docking score function has low accuracy, so a method called reranking is used to re-evaluate the docked structures. As a reranking method, a technique has been developed that uses graph neural networks (GNN) to learn about protein-protein interactions using graph structures. However, existing GNNs for reranking have a problem in that they do not fully capture three-dimensional structural features because they do not consider information about edge angles. In this study, we aimed to improve the prediction accuracy by using an architecture that considers angle information. As a result, we succeeded in achieving higher accuracy with the model that included angle information compared to the model without angle information.