O07_05
Development of Scaffold and Fragment Definition Algorithms with a Case Study on Chemical Library Analysis
Kazuma KAITOH *, Yoshihiro YAMANISHI
Graduate School of Informatics, Nagoya University
( * E-mail: kaitoh@i.nagoya-u.ac.jp )
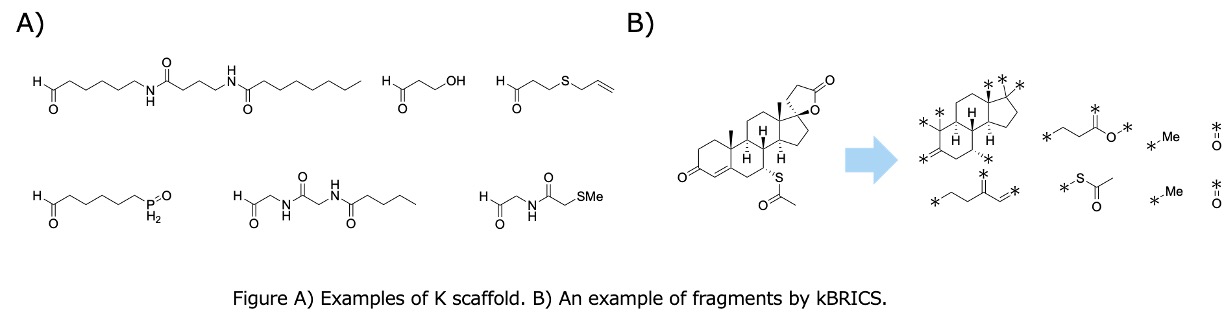
One of the key roles of chemoinformatics in drug discovery is the analysis of chemical libraries. By integrating insights from these analyses into the design of new chemical libraries, the identification of hit and lead compounds can be significantly enhanced. A common approach to analyzing chemical libraries involves dividing compounds into substructures. Scaffold extraction, which identifies the core structures of compounds, is a widely employed method. Bemis-Murcko scaffold (BM scaffold)[1] is a well-established algorithm for scaffold extraction that defines ring systems as scaffolds. However, the BM scaffold has a notable limitation: it cannot define scaffolds for compounds lacking ring structures. This limitation can result in the extraction of scaffolds that do not accurately represent the structural reality of certain compounds.
Beyond scaffold extraction, several algorithms exist for fragmenting compounds, such as the Breaking of Retrosynthetically Interesting Chemical Substructures (BRICS)[2]. However, these algorithms occasionally fail to generate appropriate fragments for specific compounds. In this study, we developed K scaffold algorithm for scaffold extraction and kBRICS algorithm for fragment extraction. These methods were applied to the ChEMBL 34 database to analyze the characteristics of the resulting scaffolds and fragments. K scaffold algorithm extends BM scaffold rules by incorporating graph structures within compounds, while kBRICS enhances BRICS by adding rules based on organic chemical reactions.
When applied to the 2,180,814 organic compounds in ChEMBL 34, BM scaffold failed to define scaffolds for 19,338 compounds, whereas the K scaffold successfully defined scaffolds for all compounds. Additionally, kBRICS generated fragments for a greater number of compounds compared to BRICS, rBRICS[3], and pBRICS[4]. Detailed analyses of these findings will be presented during the conference.
[1] Bemis, G. W.; Murcko, M. A, The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39, 2887-2893.
[2] Degen, J.; et. al., On the Art of Compiling and Using ‘Drug-Like’ Chemical Fragment Spaces. ChemMedChem 2008, 3, 1503-1507.
[3] Zhang, L.; et. al., r-BRICS – A Revised BRICS Module that Breaks Ring Structures and Carbon Chain. ChemMedChem 2023, e202300202.
[4] Vangala, S. R.; et. al., pBRICS: A Novel Fragmentation Method for Explainable Property Prediction of Drug-Like Small Molecules. J. Chem. Inf. Model. 2023, 63, 5066-5076.