O07_04
Optimization of Generator Reward Function Settings for Non-covalent KRAS Inhibitors
Casey J. GALVIN *, Masakazu ATOBE
Elix, Inc.
( * E-mail: casey.galvin@elix-inc.com)
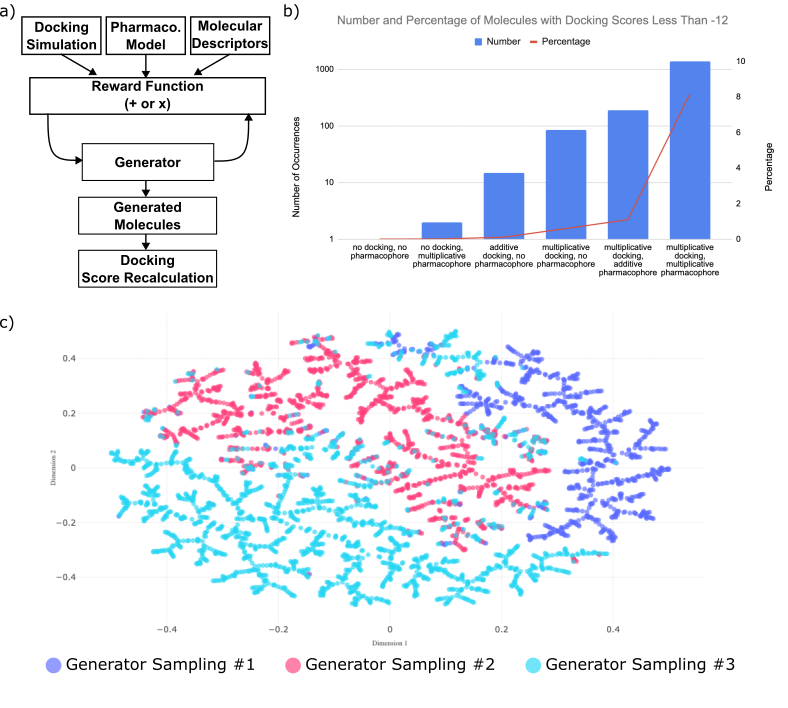
KRAS mutations are linked to various cancers, however the first inhibitors were only introduced in 2021 [1]. These inhibitors bind covalently to the cysteine residue specific to G12C, prompting the need for non-covalent inhibitors for other KRAS mutants [2]. This study combines generative models and computational chemistry techniques to design such inhibitors. We incorporate docking simulations and pharmacophore models based on a single crystal structure of a covalent KRAS inhibitor into a reward function that guides a generative model to optimize molecular design under a data-scarce regime. Incorporating either technique into the generator reward function improved the number of molecules achieving docking scores below our target threshold (lower is better), with the best results achieved by combining both pharmacophore models and docking scores as multiplicative components. This approach led to approximately 10% of the molecules (over 1,000 total) exhibiting high docking performance, which is an order of magnitude or more greater than generators without docking and/or pharmacophore model components. This finding demonstrates the strength of a multi-faceted reward function. Clustering based on molecular scaffold of high-performing molecules generated by identical reward functions run multiple times revealed distinct chemical space exploration across the different runs. This finding supports the need for multiple generator samplings to properly surface potential molecules of interest. This study highlights specific strategies in designing non-covalent pan-KRAS inhibitors by optimizing reward functions that integrate docking simulations and pharmacophore modeling. The synergy between these rewards as multiplicative factors underscores the importance of multi-component reward functions in advancing drug discovery, and identifies a strategy of overcoming the data scarcity typical of early drug discovery campaigns.
[1] Mullard, A. The KRAS Crowd Targets Its next Cancer Mutations. Nature Reviews Drug Discovery 2023, 22 (3), 167–171. https://doi.org/10.1038/d41573-023-00015-x.
[2] Kim, D.; Herdeis, L.; Rudolph, D.; Zhao, Y.; Böttcher, J.; Vides, A.; Ayala-Santos, C. I.; Pourfarjam, Y.; Cuevas-Navarro, A.; Xue, J. Y.; Mantoulidis, A.; Bröker, J.; Wunberg, T.; Schaaf, O.; Popow, J.; Wolkerstorfer, B.; Kropatsch, K. G.; Qu, R.; de Stanchina, E.; Sang, B.; Li, C.; McConnell, D. B.; Kraut, N.; Lito, P. Pan-KRAS Inhibitor Disables Oncogenic Signalling and Tumour Growth. Nature 2023, 619 (7968), 160–166. https://doi.org/10.1038/s41586-023-06123-3.